This is a preprint.
Cell morphology QTL reveal gene by environment interactions in a genetically diverse cell population
- PMID: 38014303
- PMCID: PMC10680806
- DOI: 10.1101/2023.11.18.567597
Cell morphology QTL reveal gene by environment interactions in a genetically diverse cell population
Abstract
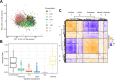
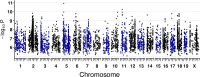
Genetically heterogenous cell lines from laboratory mice are promising tools for population-based screening as they offer power for genetic mapping, and potentially, predictive value for in vivo experimentation in genetically matched individuals. To explore this further, we derived a panel of fibroblast lines from a genetic reference population of laboratory mice (the Diversity Outbred, DO). We then used high-content imaging to capture hundreds of cell morphology traits in cells exposed to the oxidative stress-inducing arsenic metabolite monomethylarsonous acid (MMAIII). We employed dose-response modeling to capture latent parameters of response and we then used these parameters to identify several hundred cell morphology quantitative trait loci (cmQTL). Response cmQTL encompass genes with established associations with cellular responses to arsenic exposure, including Abcc4 and Txnrd1, as well as novel gene candidates like Xrcc2. Moreover, baseline trait cmQTL highlight the influence of natural variation on fundamental aspects of nuclear morphology. We show that the natural variants influencing response include both coding and non-coding variation, and that cmQTL haplotypes can be used to predict response in orthogonal cell lines. Our study sheds light on the major molecular initiating events of oxidative stress that are under genetic regulation, including the NRF2-mediated antioxidant response, cellular detoxification pathways, DNA damage repair response, and cell death trajectories.
Keywords: QTL mapping; arsenic; cell morphology; cell painting; cmQTL; fibroblasts; genetic diversity; genetic mapping; genetics; high content imaging; monomethylarsonous acid; new approach methodologies; systems genetics; systems toxicology.
Conflict of interest statement
Conflicts of Interest None to disclose.
Figures





Similar articles
-
Unraveling the genetics of arsenic toxicity with cellular morphology QTL.PLoS Genet. 2024 Apr 25;20(4):e1011248. doi: 10.1371/journal.pgen.1011248. eCollection 2024 Apr. PLoS Genet. 2024. PMID: 38662777 Free PMC article.
-
Canalization of Tomato Fruit Metabolism.Plant Cell. 2017 Nov;29(11):2753-2765. doi: 10.1105/tpc.17.00367. Epub 2017 Nov 1. Plant Cell. 2017. PMID: 29093214 Free PMC article.
-
Erratum: High-Throughput Identification of Resistance to Pseudomonas syringae pv. Tomato in Tomato using Seedling Flood Assay.J Vis Exp. 2023 Oct 18;(200). doi: 10.3791/6576. J Vis Exp. 2023. PMID: 37851522
-
Safety and nutritional assessment of GM plants and derived food and feed: the role of animal feeding trials.Food Chem Toxicol. 2008 Mar;46 Suppl 1:S2-70. doi: 10.1016/j.fct.2008.02.008. Epub 2008 Feb 13. Food Chem Toxicol. 2008. PMID: 18328408 Review.
-
QTL mapping in outbred populations: successes and challenges.Physiol Genomics. 2014 Feb 1;46(3):81-90. doi: 10.1152/physiolgenomics.00127.2013. Epub 2013 Dec 10. Physiol Genomics. 2014. PMID: 24326347 Free PMC article. Review.
References
-
- Matthew T., Jatin A., Samira A., Beth A.C., Emily P., Dhara L., Gregory W., Erin W., Aparna N., Tiffany A., et al. (2023). High-dimensional phenotyping to define the genetic basis of cellular morphology. bioRxiv, 2023.2001.2009.522731. 10.1101/2023.01.09.522731. - DOI
-
- Bray M.-A., Singh S., Han H., Davis C.T., Borgeson B., Hartland C., Kost-Alimova M., Gustafsdottir S.M., Gibson C.C., and Carpenter A.E. (2016). Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nature Protocols 11, 1757–1774. 10.1038/nprot.2016.105. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
