This is a preprint.
Expansion of human centromeric arrays in cells undergoing break-induced replication
- PMID: 38014305
- PMCID: PMC10680626
- DOI: 10.1101/2023.11.11.566714
Expansion of human centromeric arrays in cells undergoing break-induced replication
Update in
-
Expansion of human centromeric arrays in cells undergoing break-induced replication.Cell Rep. 2024 Mar 26;43(3):113851. doi: 10.1016/j.celrep.2024.113851. Epub 2024 Feb 29. Cell Rep. 2024. PMID: 38427559 Free PMC article.
Abstract
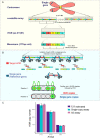
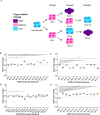
Human centromeres are located within α-satellite arrays and evolve rapidly, which can lead to individual variation in array lengths. Proposed mechanisms for such alterations in lengths are unequal cross-over between sister chromatids, gene conversion, and break-induced replication. However, the underlying molecular mechanisms responsible for the massive, complex, and homogeneous organization of centromeric arrays have not been experimentally validated. Here, we use droplet digital PCR assays to demonstrate that centromeric arrays can expand and contract within ~20 somatic cell divisions of a cell line. We find that the frequency of array variation among single-cell-derived subclones ranges from a minimum of ~7% to a maximum of ~100%. Further clonal evolution revealed that centromere expansion is favored over contraction. We find that the homologous recombination protein RAD52 and the helicase PIF1 are required for extensive array change, suggesting that centromere sequence evolution can occur via break-induced replication.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures


Similar articles
-
Expansion of human centromeric arrays in cells undergoing break-induced replication.Cell Rep. 2024 Mar 26;43(3):113851. doi: 10.1016/j.celrep.2024.113851. Epub 2024 Feb 29. Cell Rep. 2024. PMID: 38427559 Free PMC article.
-
Defining a core configuration for human centromeres during mitosis.bioRxiv [Preprint]. 2023 May 11:2023.05.10.539634. doi: 10.1101/2023.05.10.539634. bioRxiv. 2023. Update in: Nat Commun. 2023 Dec 1;14(1):7947. doi: 10.1038/s41467-023-42980-2. PMID: 37214893 Free PMC article. Updated. Preprint.
-
Protocol to measure centromeric array size changes using droplet digital PCR-based quantification of higher-order repeats.STAR Protoc. 2024 Sep 20;5(3):103218. doi: 10.1016/j.xpro.2024.103218. Epub 2024 Jul 27. STAR Protoc. 2024. PMID: 39068651 Free PMC article.
-
Structural and functional dynamics of human centromeric chromatin.Annu Rev Genomics Hum Genet. 2006;7:301-13. doi: 10.1146/annurev.genom.7.080505.115613. Annu Rev Genomics Hum Genet. 2006. PMID: 16756479 Review.
-
Plant centromere organization: a dynamic structure with conserved functions.Trends Genet. 2007 Mar;23(3):134-9. doi: 10.1016/j.tig.2007.01.004. Epub 2007 Feb 1. Trends Genet. 2007. PMID: 17275131 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
