Genotype-Phenotype Taxonomy of Hypertrophic Cardiomyopathy
- PMID: 38014537
- PMCID: PMC10729901
- DOI: 10.1161/CIRCGEN.123.004200
Genotype-Phenotype Taxonomy of Hypertrophic Cardiomyopathy
Abstract
Background: Hypertrophic cardiomyopathy (HCM) is an important cause of sudden cardiac death associated with heterogeneous phenotypes, but there is no systematic framework for classifying morphology or assessing associated risks. Here, we quantitatively survey genotype-phenotype associations in HCM to derive a data-driven taxonomy of disease expression.
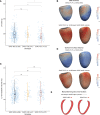
Methods: We enrolled 436 patients with HCM (median age, 60 years; 28.8% women) with clinical, genetic, and imaging data. An independent cohort of 60 patients with HCM from Singapore (median age, 59 years; 11% women) and a reference population from the UK Biobank (n=16 691; mean age, 55 years; 52.5% women) were also recruited. We used machine learning to analyze the 3-dimensional structure of the left ventricle from cardiac magnetic resonance imaging and build a tree-based classification of HCM phenotypes. Genotype and mortality risk distributions were projected on the tree.
Results: Carriers of pathogenic or likely pathogenic variants for HCM had lower left ventricular mass, but greater basal septal hypertrophy, with reduced life span (mean follow-up, 9.9 years) compared with genotype negative individuals (hazard ratio, 2.66 [95% CI, 1.42-4.96]; P<0.002). Four main phenotypic branches were identified using unsupervised learning of 3-dimensional shape: (1) nonsarcomeric hypertrophy with coexisting hypertension; (2) diffuse and basal asymmetrical hypertrophy associated with outflow tract obstruction; (3) isolated basal hypertrophy; and (4) milder nonobstructive hypertrophy enriched for familial sarcomeric HCM (odds ratio for pathogenic or likely pathogenic variants, 2.18 [95% CI, 1.93-2.28]; P=0.0001). Polygenic risk for HCM was also associated with different patterns and degrees of disease expression. The model was generalizable to an independent cohort (trustworthiness, M1: 0.86-0.88).
Conclusions: We report a data-driven taxonomy of HCM for identifying groups of patients with similar morphology while preserving a continuum of disease severity, genetic risk, and outcomes. This approach will be of value in understanding the causes and consequences of disease diversity.
Keywords: genotype; hypertension; hypertrophy; magnetic resonance imaging; phenotype.
Conflict of interest statement
Figures




References
-
- Maron BJ, Desai MY, Nishimura RA, Spirito P, Rakowski H, Towbin JA, Rowin EJ, Maron MS, Sherrid MV. Diagnosis and evaluation of hypertrophic cardiomyopathy: JACC state-of-the-art review. J Am Coll Cardiol. 2022;79:372–389. doi: 10.1016/j.jacc.2021.12.002 - PubMed
-
- Tadros R, Francis C, Xu X, Vermeer AMC, Harper AR, Huurman R, Kelu Bisabu K, Walsh R, Hoorntje ET, Te Rijdt WP, et al. . Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nat Genet. 2021;53:128–134. doi: 10.1038/s41588-020-00762-2 - PMC - PubMed
-
- Harper AR, Goel A, Grace C, Thomson KL, Petersen SE, Xu X, Waring A, Ormondroyd E, Kramer CM, Ho CY, et al. ; HCMR Investigators. Common genetic variants and modifiable risk factors underpin hypertrophic cardiomyopathy susceptibility and expressivity. Nat Genet. 2021;53:135–142. doi: 10.1038/s41588-020-00764-0 - PMC - PubMed
-
- de Marvao A, McGurk KA, Zheng SL, Thanaj M, Bai W, Duan J, Biffi C, Mazzarotto F, Statton B, Dawes TJW, et al. . Phenotypic expression and outcomes in individuals with rare genetic variants of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2021;78:1097–1110. doi: 10.1016/j.jacc.2021.07.017 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

