Altered microbiota, antimicrobial resistance genes, and functional enzyme profiles in the rumen of yak calves fed with milk replacer
- PMID: 38014976
- PMCID: PMC10871699
- DOI: 10.1128/spectrum.01314-23
Altered microbiota, antimicrobial resistance genes, and functional enzyme profiles in the rumen of yak calves fed with milk replacer
Abstract
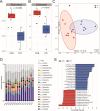
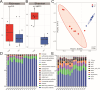
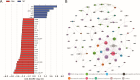
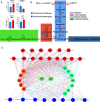
Yaks, as ruminants inhabiting high-altitude environments, possess a distinct rumen microbiome and are resistant to extreme living conditions. This study investigated the microbiota, resistome, and functional gene profiles in the rumen of yaks fed milk or milk replacer (MR), providing insights into the regulation of the rumen microbiome and the intervention of antimicrobial resistance in yaks through dietary methods. The abundance of Prevotella members increased significantly in response to MR. Tetracycline resistance was the most predominant. The rumen of yaks contained multiple antimicrobial resistance genes (ARGs) originating from different bacteria, which could be driven by MR, and these ARGs displayed intricate and complex interactions. MR also induced changes in functional genes. The enzymes associated with fiber degradation and butyrate metabolism were activated and showed close correlations with Prevotella members and butyrate concentration. This study allows us to deeply understand the ruminal microbiome and ARGs of yaks and their relationship with rumen bacteria in response to different milk sources.
Keywords: metagenomics; microbiome; milk replacer; resistome; rumen; yak.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Feeding systems influence the rumen resistome in yaks by changing the microbiome.Front Microbiol. 2025 Mar 19;16:1505938. doi: 10.3389/fmicb.2025.1505938. eCollection 2025. Front Microbiol. 2025. PMID: 40177486 Free PMC article.
-
Metagenomics reveals the temporal dynamics of the rumen resistome and microbiome in goat kids.Microbiome. 2024 Jan 22;12(1):14. doi: 10.1186/s40168-023-01733-5. Microbiome. 2024. PMID: 38254181 Free PMC article.
-
Ruminal resistome of dairy cattle is individualized and the resistotypes are associated with milking traits.Anim Microbiome. 2021 Feb 10;3(1):18. doi: 10.1186/s42523-021-00081-9. Anim Microbiome. 2021. PMID: 33568223 Free PMC article.
-
Linkages between rumen microbiome, host, and environment in yaks, and their implications for understanding animal production and management.Front Microbiol. 2024 Jan 29;15:1301258. doi: 10.3389/fmicb.2024.1301258. eCollection 2024. Front Microbiol. 2024. PMID: 38348184 Free PMC article. Review.
-
Review: Exogenous butyrate: implications for the functional development of ruminal epithelium and calf performance.Animal. 2017 Sep;11(9):1522-1530. doi: 10.1017/S1751731117000167. Epub 2017 Feb 14. Animal. 2017. PMID: 28193308 Review.
Cited by
-
Core microbe Bifidobacterium in the hindgut of calves improves the growth phenotype of young hosts by regulating microbial functions and host metabolism.Microbiome. 2025 Jan 16;13(1):13. doi: 10.1186/s40168-024-02010-9. Microbiome. 2025. PMID: 39819813 Free PMC article.
-
Spatio-temporal characteristics of the gastrointestinal resistome in a cow-to-calf model and its environmental dissemination in a dairy production system.Imeta. 2025 May 14;4(4):e70047. doi: 10.1002/imt2.70047. eCollection 2025 Aug. Imeta. 2025. PMID: 40860434 Free PMC article.
-
Maternal gastrointestinal microbiome shapes gut microbial function and resistome of newborns in a cow-to-calf model.Microbiome. 2024 Oct 22;12(1):216. doi: 10.1186/s40168-024-01943-5. Microbiome. 2024. PMID: 39438998 Free PMC article.
-
Exploration of mobile genetic elements in the ruminal microbiome of Nellore cattle.Sci Rep. 2024 Jun 6;14(1):13056. doi: 10.1038/s41598-024-63951-7. Sci Rep. 2024. PMID: 38844487 Free PMC article.
-
Seasonal stability of the rumen microbiome contributes to the adaptation patterns to extreme environmental conditions in grazing yak and cattle.BMC Biol. 2024 Oct 23;22(1):240. doi: 10.1186/s12915-024-02035-4. BMC Biol. 2024. PMID: 39443951 Free PMC article.
References
-
- Cui Z, Wu S, Liu S, Sun L, Feng Y, Cao Y, Chai S, Zhang G, Yao J. 2020. From maternal grazing to barn feeding during pre-weaning period: altered gastrointestinal microbiota contributes to change the development and function of the rumen and intestine of yak calves. Front Microbiol 11:485. doi:10.3389/fmicb.2020.00485 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

