Comparing Microsporidia-targeting primers for environmental DNA sequencing
- PMID: 38015008
- PMCID: PMC10683580
- DOI: 10.1051/parasite/2023056
Comparing Microsporidia-targeting primers for environmental DNA sequencing
Abstract
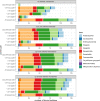
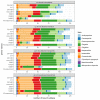
Metabarcoding is a powerful tool to detect classical, and well-known "long-branch" Microsporidia in environmental samples. Several primer pairs were developed to target these unique microbial parasites, the majority of which remain undetected when using general metabarcoding primers. Most of these Microsporidia-targeting primer pairs amplify fragments of different length of the small subunit ribosomal RNA (SSU-rRNA) gene. However, we lack a broad comparison of the efficacy of those primers. Here, we conducted in silico PCRs with three short-read (which amplify a few-hundred base pairs) and two long-read (which amplify over a thousand base pairs) metabarcoding primer pairs on a variety of publicly available Microsporidia sensu lato SSU-rRNA gene sequences to test which primers capture most of the Microsporidia diversity. Our results indicate that the primer pairs do result in slight differences in inferred richness. Furthermore, some of the reverse primers are also able to bind to microsporidian subtaxa beyond the classical Microsporidia, which include the metchnikovellidan Amphiamblys spp., the chytridiopsid Chytridiopsis typographi and the "short-branch" microsporidian Mitosporidium daphniae.
Title: Comparaison des amorces ciblant les Microsporidies pour le séquençage de l’ADN environnemental.
Abstract: Le métabarcoding est un outil puissant pour détecter les microsporidies classiques et bien connues à « longues branches » dans les échantillons environnementaux. Plusieurs paires d’amorces ont été développées pour cibler ces parasites microscopiques exceptionnels, dont la majorité restent indétectables lors de l’utilisation d’amorces générales de métabarcoding. La plupart de ces paires d’amorces ciblant les microsporidies amplifient des fragments de différentes longueurs du gène de la petite sous-unité de l’ARN ribosomal (SSU-rRNA). Cependant, nous manquons d’une comparaison générale de l’efficacité de ces amorces. Ici, pour tester quelles amorces capturent la plus grande partie de la diversité des microsporidies, nous avons réalisé des PCR in silico avec trois paires d’amorces de métabarcoding à lecture courte (qui amplifient quelques centaines de paires de bases) et deux paires d’amorces de métabarcoding à lecture longue (qui amplifient plus d’un millier de bases), sur une variété de séquences du gène SSU-rRNA de Microsporidia sensu lato accessibles au public. Nos résultats indiquent que les paires d’amorces entraînent de légères différences dans la richesse déduite. En outre, certaines des amorces inverses sont également capables de se lier à des sous-taxons de microsporidies au-delà des Microsporidia classiques, notamment les Metchnikovellidae Amphiamblys spp., le Chytridiopsida Chytridiopsis typographi et la microsporidie à « branches courtes » Mitosporidium daphniae.
Keywords: Barcoding; Diversity; Metabarcoding; Microsporidia; Parasites; Protists.
© A. Doliwa et al., published by EDP Sciences, 2023.
Figures
References
-
- Ardila-Garcia AM, Raghuram N, Sihota P, Fast NM. 2013. Microsporidian diversity in soil, sand, and compost of the Pacific Northwest. Journal of Eukaryotic Microbiology, 60, 601–608. - PubMed
-
- Baker MD, Vossbrinck CR, Didier ES, Maddox JV, Shadduck JA. 1995. Small subunit ribosomal DNA phylogeny of various microsporidia with emphasis on AIDS related forms. Journal of Eukaryotic Microbiology, 42, 564–570. - PubMed
-
- Bass D, Christison KW, Stentiford GD, Cook LSJ, Hartikainen H. 2023. Environmental DNA/RNA for pathogen and parasite detection, surveillance, and ecology. Trends in Parasitology, 39, 285–304. - PubMed
-
- Bojko J, Reinke AW, Stentiford GD, Williams B, Rogers MSJ, Bass D. 2022. Microsporidia: a new taxonomic, evolutionary, and ecological synthesis. Trends in Parasitology, 38, 642–659. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
