Internal RNA 2'-O-methylation on the HIV-1 genome impairs reverse transcription
- PMID: 38015463
- PMCID: PMC10853786
- DOI: 10.1093/nar/gkad1134
Internal RNA 2'-O-methylation on the HIV-1 genome impairs reverse transcription
Abstract
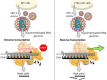
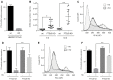
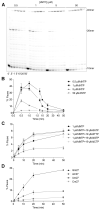
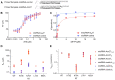
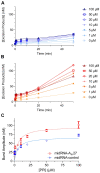
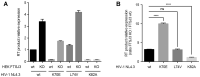
Viral RNA genomes are modified by epitranscriptomic marks, including 2'-O-methylation that is added by cellular or viral methyltransferases. 2'-O-Methylation modulates RNA structure, function and discrimination between self- and non-self-RNA by innate immune sensors such as RIG-I-like receptors. This is illustrated by human immunodeficiency virus type-1 (HIV-1) that decorates its RNA genome through hijacking the cellular FTSJ3 2'-O-methyltransferase, thereby limiting immune sensing and interferon production. However, the impact of such an RNA modification during viral genome replication is poorly understood. Here we show by performing endogenous reverse transcription on methylated or hypomethylated HIV-1 particles, that 2'-O-methylation negatively affects HIV-1 reverse transcriptase activity. Biochemical assays confirm that RNA 2'-O-methylation impedes reverse transcriptase activity, especially at low dNTP concentrations reflecting those in quiescent cells, by reducing nucleotide incorporation efficiency and impairing translocation. Mutagenesis highlights K70 as a critical amino acid for the reverse transcriptase to bypass 2'-O-methylation. Hence, the observed antiviral effect due to viral RNA 2'-O-methylation antagonizes the FTSJ3-mediated proviral effects, suggesting the fine-tuning of RNA methylation during viral replication.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

