Nucleolin binds to and regulates transcription of hepatitis B virus covalently closed circular DNA minichromosome
- PMID: 38015841
- PMCID: PMC10710063
- DOI: 10.1073/pnas.2306390120
Nucleolin binds to and regulates transcription of hepatitis B virus covalently closed circular DNA minichromosome
Abstract
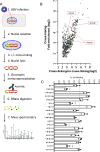
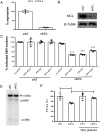
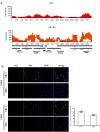
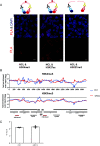
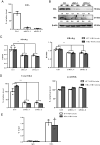
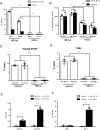
Hepatitis B virus (HBV) remains a major public health threat with nearly 300 million people chronically infected worldwide who are at a high risk of developing hepatocellular carcinoma. Current therapies are effective in suppressing HBV replication but rarely lead to cure. Current therapies do not affect the HBV covalently closed circular DNA (cccDNA), which serves as the template for viral transcription and replication and is highly stable in infected cells to ensure viral persistence. In this study, we aim to identify and elucidate the functional role of cccDNA-associated host factors using affinity purification and protein mass spectrometry in HBV-infected cells. Nucleolin was identified as a key cccDNA-binding protein and shown to play an important role in HBV cccDNA transcription, likely via epigenetic regulation. Targeting nucleolin to silence cccDNA transcription in infected hepatocytes may be a promising therapeutic strategy for a functional cure of HBV.
Keywords: HBV minichromosome; antiviral; epigenetics; proteomics; viral replication.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

