The USP7-STAT3-granzyme-Par-1 axis regulates allergic inflammation by promoting differentiation of IL-5-producing Th2 cells
- PMID: 38015852
- PMCID: PMC10710068
- DOI: 10.1073/pnas.2302903120
The USP7-STAT3-granzyme-Par-1 axis regulates allergic inflammation by promoting differentiation of IL-5-producing Th2 cells
Abstract
Uncontrolled type 2 immunity by type 2 helper T (Th2) cells causes intractable allergic diseases; however, whether the interaction of CD4+ T cells shapes the pathophysiology of allergic diseases remains unclear. We identified a subset of Th2 cells that produced the serine proteases granzyme A and B early in differentiation. Granzymes cleave protease-activated receptor (Par)-1 and induce phosphorylation of p38 mitogen-activated protein kinase (MAPK), resulting in the enhanced production of IL-5 and IL-13 in both mouse and human Th2 cells. Ubiquitin-specific protease 7 (USP7) regulates IL-4-induced phosphorylation of STAT3, resulting in granzyme production during Th2 cell differentiation. Genetic deletion of Usp7 or Gzma and pharmacological blockade of granzyme B ameliorated allergic airway inflammation. Furthermore, PAR-1+ and granzyme+ Th2 cells were colocalized in nasal polyps from patients with eosinophilic chronic rhinosinusitis. Thus, the USP7-STAT3-granzymes-Par-1 pathway is a potential therapeutic target for intractable allergic diseases.
Keywords: asthma, intractable allergic disease; granzyme; pathogenic Th2 cell; signal transducers and activators of transcription (STAT)3; ubiquitin specific protease 7 (USP7).
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

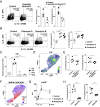

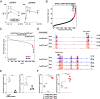

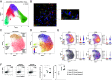
References
-
- Nakayama T., et al. , Th2 cells in health and disease. Annu. Rev. Immunol. 35, 53–84 (2017). - PubMed
-
- Ruterbusch M., Pruner K. B., Shehata L., Pepper M., In vivo CD4(+) T cell differentiation and function: Revisiting the Th1/Th2 paradigm. Annu. Rev. Immunol. 38, 705–725 (2020). - PubMed
-
- Cabeza-Cabrerizo M., Cardoso A., Minutti C. M., Pereira da Costa M., Reis e Sousa C., Dendritic cells revisited. Annu. Rev. Immunol. 39, 131–166 (2021). - PubMed
-
- Tibbitt C. A., et al. , Single-cell RNA sequencing of the T helper cell response to house dust mites defines a distinct gene expression signature in airway Th2 cells. Immunity 51, 169–184.e165 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- JP19H05650/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- 20H03685/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- 17K08876/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- 18K07164/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- 19K16683/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- 21H05121/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- 19K23858/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- 22K15485/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- JP21H05120/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- JP20ek0410060/Japan Agency for Medical Research and Development (AMED)
- JP22ek0410092/Japan Agency for Medical Research and Development (AMED)
- JP20gm1210003/Japan Agency for Medical Research and Development (AMED)
- JPMJFR200R/JST FORREST program
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

