Identification of the consistently differential expressed hub mRNAs and proteins in lung adenocarcinoma and construction of the prognostic signature: a multidimensional analysis
- PMID: 38016140
- PMCID: PMC10871637
- DOI: 10.1097/JS9.0000000000000943
Identification of the consistently differential expressed hub mRNAs and proteins in lung adenocarcinoma and construction of the prognostic signature: a multidimensional analysis
Abstract
Background: This study aimed to elucidate the consistency of differentially expressed hub mRNAs and proteins in lung adenocarcinoma (LUAD) across populations and to construct a comprehensive LUAD prognostic signature.
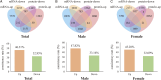
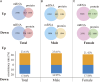
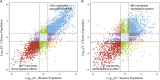
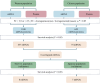
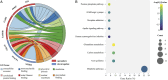
Methods: The transcriptomic and proteomics data from different populations were standardized and analyzed using the same criteria to identify the consistently differential expressed mRNAs and proteins across genders and races. We then integrated prognosis-related mRNAs with clinical, pathological, and EGFR (epidermal growth factor receptor) mutation data to construct a survival model, subsequently validating it across populations. Through plasma proteomics, plasma proteins that consistently differential expressed with LUAD tissues were screened and validated, with their associations discerned by measuring expressions in tumor tissues and tumor vascular normalization.
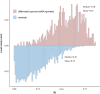
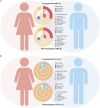
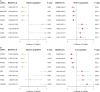
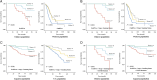
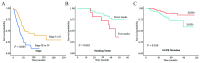
Results: The consistency rate of differentially expressed mRNAs and proteins was ~20-40%, with ethnic factors leading to about 40-60% consistency of differentially expressed mRNA or protein across populations. The survival model based on the identified eight hub mRNAs as well as stage, smoking status, and EGFR mutations, demonstrated good prognostic prediction capabilities in both Western and East Asian populations, with a higher number of unfavorable variables indicating poorer LUAD prognosis. Notably, GPI expression in tumor tissues was inversely correlated with vascular normalization and positively correlated with plasma GPI expression.
Conclusion: Our study underscores the significance of integrating transcriptomics and proteomics data, emphasizing the need to account for genetic diversity among ethnic groups. The developed survival model may offer a holistic perspective on LUAD progression, enhancing prognosis and therapeutic strategies.
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Figures


References
-
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17–48. - PubMed
-
- Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc 2019;94:1623–1640. - PubMed
-
- Yu XQ, Yap ML, Cheng ES, et al. Evaluating prognostic factors for sex differences in lung cancer survival: findings from a large Australian cohort. J Thorac Oncol 2022;17:688–699. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

