Multimodal subspace identification for modeling discrete-continuous spiking and field potential population activity
- PMID: 38016450
- PMCID: PMC10913727
- DOI: 10.1088/1741-2552/ad1053
Multimodal subspace identification for modeling discrete-continuous spiking and field potential population activity
Abstract
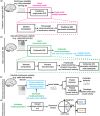
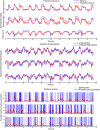
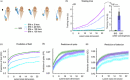
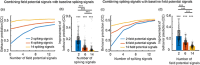
Objective.Learning dynamical latent state models for multimodal spiking and field potential activity can reveal their collective low-dimensional dynamics and enable better decoding of behavior through multimodal fusion. Toward this goal, developing unsupervised learning methods that are computationally efficient is important, especially for real-time learning applications such as brain-machine interfaces (BMIs). However, efficient learning remains elusive for multimodal spike-field data due to their heterogeneous discrete-continuous distributions and different timescales.Approach.Here, we develop a multiscale subspace identification (multiscale SID) algorithm that enables computationally efficient learning for modeling and dimensionality reduction for multimodal discrete-continuous spike-field data. We describe the spike-field activity as combined Poisson and Gaussian observations, for which we derive a new analytical SID method. Importantly, we also introduce a novel constrained optimization approach to learn valid noise statistics, which is critical for multimodal statistical inference of the latent state, neural activity, and behavior. We validate the method using numerical simulations and with spiking and local field potential population activity recorded during a naturalistic reach and grasp behavior.Main results.We find that multiscale SID accurately learned dynamical models of spike-field signals and extracted low-dimensional dynamics from these multimodal signals. Further, it fused multimodal information, thus better identifying the dynamical modes and predicting behavior compared to using a single modality. Finally, compared to existing multiscale expectation-maximization learning for Poisson-Gaussian observations, multiscale SID had a much lower training time while being better in identifying the dynamical modes and having a better or similar accuracy in predicting neural activity and behavior.Significance.Overall, multiscale SID is an accurate learning method that is particularly beneficial when efficient learning is of interest, such as for online adaptive BMIs to track non-stationary dynamics or for reducing offline training time in neuroscience investigations.
Keywords: Poisson and Gaussian data; dynamical systems; local field potentials; multimodal data; spiking activity; unsupervised learning.
Creative Commons Attribution license.
Figures




Update of
-
Multimodal subspace identification for modeling discrete-continuous spiking and field potential population activity.bioRxiv [Preprint]. 2023 May 30:2023.05.26.542509. doi: 10.1101/2023.05.26.542509. bioRxiv. 2023. Update in: J Neural Eng. 2024 Mar 01;21(2). doi: 10.1088/1741-2552/ad1053. PMID: 37398400 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
