TScan-II: A genome-scale platform for the de novo identification of CD4+ T cell epitopes
- PMID: 38016469
- PMCID: PMC10841602
- DOI: 10.1016/j.cell.2023.10.024
TScan-II: A genome-scale platform for the de novo identification of CD4+ T cell epitopes
Abstract
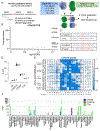
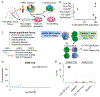
CD4+ T cells play fundamental roles in orchestrating immune responses and tissue homeostasis. However, our inability to associate peptide human leukocyte antigen class-II (HLA-II) complexes with their cognate T cell receptors (TCRs) in an unbiased manner has hampered our understanding of CD4+ T cell function and role in pathologies. Here, we introduce TScan-II, a highly sensitive genome-scale CD4+ antigen discovery platform. This platform seamlessly integrates the endogenous HLA-II antigen-processing machinery in synthetic antigen-presenting cells and TCR signaling in T cells, enabling the simultaneous screening of multiple HLAs and TCRs. Leveraging genome-scale human, virome, and epitope mutagenesis libraries, TScan-II facilitates de novo antigen discovery and deep exploration of TCR specificity. We demonstrate TScan-II's potential for basic and translational research by identifying a non-canonical antigen for a cancer-reactive CD4+ T cell clone. Additionally, we identified two antigens for clonally expanded CD4+ T cells in Sjögren's disease, which bind distinct HLAs and are expressed in HLA-II-positive ductal cells within affected salivary glands.
Keywords: CD4+ T cell; HIV; Pancreatic cancer; Sjögren’s disease; T cell epitope; T cell screening; antigen discovery; antigen library; epitope discovery; epitope mutagenesis.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests M.H.D., T.K., and S.J.E. are patent holders of the TScan-I and TScan-II technologies. T.K. is a co-founder of TScan Therapeutics and ImmuneID and serves on the Scientific Advisory Board of TScan Therapeutics. S.J.E. is a founder of TScan Therapeutics, ImmuneID, MAZE Therapeutics, and Mirimus. S.J.E. serves on the Scientific Advisory Board of TScan Therapeutics and Maze Therapeutics. In accordance with Partners HealthCare's conflict of interest policies, the Partners Office for Interactions with Industry has reviewed S.J.E.’s financial interest in TScan and determined that it creates no significant risk to the welfare of the participants in this study or to the integrity of the research.
Figures





Comment on
-
Re-centauring T cell antigen discovery around CD4+ T cells.Cell Rep Methods. 2024 Jan 22;4(1):100694. doi: 10.1016/j.crmeth.2023.100694. Cell Rep Methods. 2024. PMID: 38262348 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

