CerS6-dependent ceramide synthesis in hypothalamic neurons promotes ER/mitochondrial stress and impairs glucose homeostasis in obese mice
- PMID: 38016943
- PMCID: PMC10684560
- DOI: 10.1038/s41467-023-42595-7
CerS6-dependent ceramide synthesis in hypothalamic neurons promotes ER/mitochondrial stress and impairs glucose homeostasis in obese mice
Abstract
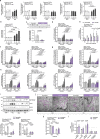
Dysregulation of hypothalamic ceramides has been associated with disrupted neuronal pathways in control of energy and glucose homeostasis. However, the specific ceramide species promoting neuronal lipotoxicity in obesity have remained obscure. Here, we find increased expression of the C16:0 ceramide-producing ceramide synthase (CerS)6 in cultured hypothalamic neurons exposed to palmitate in vitro and in the hypothalamus of obese mice. Conditional deletion of CerS6 in hypothalamic neurons attenuates high-fat diet (HFD)-dependent weight gain and improves glucose metabolism. Specifically, CerS6 deficiency in neurons expressing pro-opiomelanocortin (POMC) or steroidogenic factor 1 (SF-1) alters feeding behavior and alleviates the adverse metabolic effects of HFD feeding on insulin sensitivity and glucose tolerance. POMC-expressing cell-selective deletion of CerS6 prevents the diet-induced alterations of mitochondrial morphology and improves cellular leptin sensitivity. Our experiments reveal functions of CerS6-derived ceramides in hypothalamic lipotoxicity, altered mitochondrial dynamics, and ER/mitochondrial stress in the deregulation of food intake and glucose metabolism in obesity.
© 2023. The Author(s).
Conflict of interest statement
J.C.B. is co-founder of Cerapeutix and has received research funding through collaborations with Sanofi Aventis and Novo Nordisk Inc., which did not affect the content of this article. The remaining authors declare no competing interests.
Figures

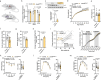



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

