Nuclear actin polymerization rapidly mediates replication fork remodeling upon stress by limiting PrimPol activity
- PMID: 38016948
- PMCID: PMC10684888
- DOI: 10.1038/s41467-023-43183-5
Nuclear actin polymerization rapidly mediates replication fork remodeling upon stress by limiting PrimPol activity
Abstract
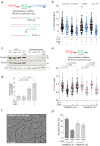
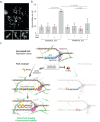
Cells rapidly respond to replication stress actively slowing fork progression and inducing fork reversal. How replication fork plasticity is achieved in the context of nuclear organization is currently unknown. Using nuclear actin probes in living and fixed cells, we visualized nuclear actin filaments in unperturbed S phase and observed their rapid extension in number and length upon genotoxic treatments, frequently taking contact with replication factories. Chemically or genetically impairing nuclear actin polymerization shortly before these treatments prevents active fork slowing and abolishes fork reversal. Defective fork remodeling is linked to deregulated chromatin loading of PrimPol, which promotes unrestrained and discontinuous DNA synthesis and limits the recruitment of RAD51 and SMARCAL1 to nascent DNA. Moreover, defective nuclear actin polymerization upon mild replication interference induces chromosomal instability in a PRIMPOL-dependent manner. Hence, by limiting PrimPol activity, nuclear F-actin orchestrates replication fork plasticity and is a key molecular determinant in the rapid cellular response to genotoxic treatments.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Update of
-
Replication fork plasticity upon replication stress requires rapid nuclear actin polymerization.bioRxiv [Preprint]. 2023 Mar 25:2023.03.24.534097. doi: 10.1101/2023.03.24.534097. bioRxiv. 2023. Update in: Nat Commun. 2023 Nov 28;14(1):7819. doi: 10.1038/s41467-023-43183-5. PMID: 36993227 Free PMC article. Updated. Preprint.
References
-
- Berti M, Cortez D, Lopes M. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell Biol. 2020;18:622–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

