Single cell and spatial transcriptomics analysis of kidney double negative T lymphocytes in normal and ischemic mouse kidneys
- PMID: 38017015
- PMCID: PMC10684868
- DOI: 10.1038/s41598-023-48213-2
Single cell and spatial transcriptomics analysis of kidney double negative T lymphocytes in normal and ischemic mouse kidneys
Abstract
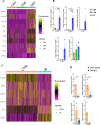
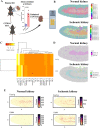
T cells are important in the pathogenesis of acute kidney injury (AKI), and TCR+CD4-CD8- (double negative-DN) are T cells that have regulatory properties. However, there is limited information on DN T cells compared to traditional CD4+ and CD8+ cells. To elucidate the molecular signature and spatial dynamics of DN T cells during AKI, we performed single-cell RNA sequencing (scRNA-seq) on sorted murine DN, CD4+, and CD8+ cells combined with spatial transcriptomic profiling of normal and post AKI mouse kidneys. scRNA-seq revealed distinct transcriptional profiles for DN, CD4+, and CD8+ T cells of mouse kidneys with enrichment of Kcnq5, Klrb1c, Fcer1g, and Klre1 expression in DN T cells compared to CD4+ and CD8+ T cells in normal kidney tissue. We validated the expression of these four genes in mouse kidney DN, CD4+ and CD8+ T cells using RT-PCR and Kcnq5, Klrb1, and Fcer1g genes with the NIH human kidney precision medicine project (KPMP). Spatial transcriptomics in normal and ischemic mouse kidney tissue showed a localized cluster of T cells in the outer medulla expressing DN T cell genes including Fcer1g. These results provide a template for future studies in DN T as well as CD4+ and CD8+ cells in normal and diseased kidneys.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

