Chronic lymphocytic leukemia patient-derived xenografts recapitulate clonal evolution to Richter transformation
- PMID: 38017105
- PMCID: PMC10912031
- DOI: 10.1038/s41375-023-02095-5
Chronic lymphocytic leukemia patient-derived xenografts recapitulate clonal evolution to Richter transformation
Abstract
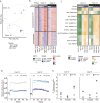
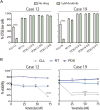
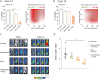
Chronic lymphocytic leukemia (CLL) is a B-cell neoplasm with a heterogeneous clinical behavior. In 5-10% of patients the disease transforms into a diffuse large-B cell lymphoma known as Richter transformation (RT), which is associated with dismal prognosis. Here, we aimed to establish patient-derived xenograft (PDX) models to study the molecular features and evolution of CLL and RT. We generated two PDXs by injecting CLL (PDX12) and RT (PDX19) cells into immunocompromised NSG mice. Both PDXs were morphologically and phenotypically similar to RT. Whole-genome sequencing analysis at different time points of the PDX evolution revealed a genomic landscape similar to RT tumors from both patients and uncovered an unprecedented RT subclonal heterogeneity and clonal evolution during PDX generation. In PDX12, the transformed cells expanded from a very small subclone already present at the CLL stage. Transcriptomic analysis of PDXs showed a high oxidative phosphorylation (OXPHOS) and low B-cell receptor (BCR) signaling similar to the RT in the patients. IACS-010759, an OXPHOS inhibitor, reduced proliferation, and circumvented resistance to venetoclax. In summary, we have generated new RT-PDX models, one of them from CLL cells that mimicked the evolution of CLL to RT uncovering intrinsic features of RT cells of therapeutical value.
© 2023. The Author(s).
Conflict of interest statement
E.C. has been a consultant for Takeda, NanoString, AbbVie and Illumina; has received honoraria from Janssen, EUSPharma and Roche for speaking at educational activities and research funding from AstraZeneca and is an inventor on 2 patents filed by the National Institutes of Health, National Cancer Institute: “Methods for selecting and treating lymphoma types,” licensed to NanoString Technologies, and “Evaluation of mantle cell lymphoma and methods related thereof”, not related to this project. F.N. has received honoraria from Janssen, AbbVie, AstraZeneca and Sophia Genetics for speaking at educational activities. D.C. has received honoraria from AbbVie, Sophia Genetics, Thermofisher and AstraZeneca for speaking at educational activities. The remaining authors declare no competing financial interests.
Figures




Similar articles
-
Mouse models of chronic lymphocytic leukemia and Richter transformation: what we have learnt and what we are missing.Front Immunol. 2024 Jun 6;15:1376660. doi: 10.3389/fimmu.2024.1376660. eCollection 2024. Front Immunol. 2024. PMID: 38903501 Free PMC article. Review.
-
PD-1 Expression in Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma (CLL/SLL) and Large B-cell Richter Transformation (DLBCL-RT): A Characteristic Feature of DLBCL-RT and Potential Surrogate Marker for Clonal Relatedness.Am J Surg Pathol. 2018 Jul;42(7):843-854. doi: 10.1097/PAS.0000000000001077. Am J Surg Pathol. 2018. PMID: 29762141
-
Detection of early seeding of Richter transformation in chronic lymphocytic leukemia.Nat Med. 2022 Aug;28(8):1662-1671. doi: 10.1038/s41591-022-01927-8. Epub 2022 Aug 11. Nat Med. 2022. PMID: 35953718 Free PMC article.
-
Descriptive analysis of genetic aberrations and cell of origin in Richter transformation.Leuk Lymphoma. 2019 Apr;60(4):971-979. doi: 10.1080/10428194.2018.1516878. Epub 2019 Jan 11. Leuk Lymphoma. 2019. PMID: 30632835
-
Clonal relatedness as a prognostic marker in Richter transformation of chronic lymphocytic leukemia: a systematic review.Blood Adv. 2025 Jun 10;9(11):2773-2780. doi: 10.1182/bloodadvances.2024015594. Blood Adv. 2025. PMID: 40127387 Free PMC article.
Cited by
-
Mouse models of chronic lymphocytic leukemia and Richter transformation: what we have learnt and what we are missing.Front Immunol. 2024 Jun 6;15:1376660. doi: 10.3389/fimmu.2024.1376660. eCollection 2024. Front Immunol. 2024. PMID: 38903501 Free PMC article. Review.
-
[Establishment of a chronic lymphocytic leukemia mouse model via adoptive transfer of Eμ-TCL1 transgenic splenocytes].Zhonghua Xue Ye Xue Za Zhi. 2025 May 14;46(5):445-452. doi: 10.3760/cma.j.cn121090-20241120-00461. Zhonghua Xue Ye Xue Za Zhi. 2025. PMID: 40623904 Free PMC article. Chinese.
-
A question of TiME: how microenvironmental interactions shape response to immunotherapy in CLL and Richter Transformation.Front Immunol. 2025 May 29;16:1592574. doi: 10.3389/fimmu.2025.1592574. eCollection 2025. Front Immunol. 2025. PMID: 40510357 Free PMC article. Review.
-
Can O-GIcNAc Transferase (OGT) Complex Be Used as a Target for the Treatment of Hematological Malignancies?Pharmaceuticals (Basel). 2024 May 22;17(6):664. doi: 10.3390/ph17060664. Pharmaceuticals (Basel). 2024. PMID: 38931332 Free PMC article. Review.
References
-
- Nadeu F, Diaz-Navarro A, Delgado J, Puente XS, Campo E. Genomic and epigenomic alterations in chronic lymphocytic leukemia. Annu Rev Pathol: Mech Dis. 2020;15:149–77. - PubMed
-
- Kulis M, Heath S, Bibikova M, Queirós AC, Navarro A, Clot G, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–42. - PubMed
-
- Puente XS, Beà S, Valdés-Mas R, Villamor N, Gutiérrez-Abril J, Martín-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24. - PubMed
Publication types
MeSH terms
Grants and funding
- RTI2018-094584-B-I00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- PID2021-123165OB-I00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- 21-40-11-NADE/American Association for Cancer Research (American Association for Cancer Research, Inc.)
- RG-202012-00245/European Hematology Association (EHA)
- LADY_TATA_21_3223/Lady Tata Memorial Trust
LinkOut - more resources
Full Text Sources

