On the origin of the nucleus: a hypothesis
- PMID: 38018971
- PMCID: PMC10732040
- DOI: 10.1128/mmbr.00186-21
On the origin of the nucleus: a hypothesis
Abstract
SUMMARYIn this hypothesis article, we explore the origin of the eukaryotic nucleus. In doing so, we first look afresh at the nature of this defining feature of the eukaryotic cell and its core functions-emphasizing the utility of seeing the eukaryotic nucleoplasm and cytoplasm as distinct regions of a common compartment. We then discuss recent progress in understanding the evolution of the eukaryotic cell from archaeal and bacterial ancestors, focusing on phylogenetic and experimental data which have revealed that many eukaryotic machines with nuclear activities have archaeal counterparts. In addition, we review the literature describing the cell biology of representatives of the TACK and Asgardarchaeaota - the closest known living archaeal relatives of eukaryotes. Finally, bringing these strands together, we propose a model for the archaeal origin of the nucleus that explains much of the current data, including predictions that can be used to put the model to the test.
Keywords: archaea; asgard archaea; eukaryogenesis; evolution; inside-out model; nucleolus; nucleus; origins; theory; topology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

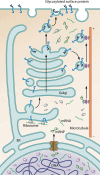



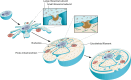
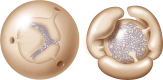
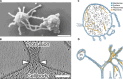
References
-
- Heimerl T, Flechsler J, Pickl C, Heinz V, Salecker B, Zweck J, Wanner G, Geimer S, Samson RY, Bell SD, Huber H, Wirth R, Wurch L, Podar M, Rachel R. 2017. A complex endomembrane system in the archaeon Ignicoccus hospitalis tapped by Nanoarchaeum equitans. Front Microbiol 8:1072. doi:10.3389/fmicb.2017.01072 - DOI - PMC - PubMed
-
- Volland J-M, Gonzalez-Rizzo S, Gros O, Tyml T, Ivanova N, Schulz F, Goudeau D, Elisabeth NH, Nath N, Udwary D, Malmstrom RR, Guidi-Rontani C, Bolte-Kluge S, Davies KM, Jean MR, Mansot J-L, Mouncey NJ, Angert ER, Woyke T, Date SV. 2022. A centimeter-long bacterium with DNA contained in metabolically active, membrane-bound organelles. Science 376:1453–1458. doi:10.1126/science.abb3634 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

