Cryptic-site-specific antibodies to the SARS-CoV-2 receptor binding domain can retain functional binding affinity to spike variants
- PMID: 38019013
- PMCID: PMC10746274
- DOI: 10.1128/jvi.01070-23
Cryptic-site-specific antibodies to the SARS-CoV-2 receptor binding domain can retain functional binding affinity to spike variants
Abstract
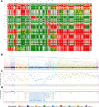
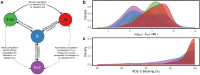
Multiple SARS-CoV-2 variants of concern have emerged and caused a significant number of infections and deaths worldwide. These variants of concern contain mutations that might significantly affect antigen-targeting by antibodies. It is therefore important to further understand how antibody binding and neutralization are affected by the mutations in SARS-CoV-2 variants. We highlighted how antibody epitope specificity can influence antibody binding to SARS-CoV-2 spike protein variants and neutralization of SARS-CoV-2 variants. We showed that weakened spike binding and neutralization of Beta (B.1.351) and Omicron (BA.1) variants compared to wildtype are not universal among the panel of antibodies and identified antibodies of a specific binding footprint exhibiting consistent enhancement of spike binding and retained neutralization to Beta variant. These data and analysis can inform how antigen-targeting by antibodies might evolve during a pandemic and prepare for potential future sarbecovirus outbreaks.
Keywords: ACE-2 blocking; COVID-19; RBD; SARS-CoV-2; binding kinetics; biolayer interferometry; epitope binning; monoclonal antibodies; neutralizing antibodies; surface plasmon resonance.
Conflict of interest statement
Daniel Bedinger is an employee of Carterra, Inc., and has stock options.
Figures




References
-
- Costanzo M, De Giglio MAR, Roviello GN. 2022. Anti-coronavirus vaccines: past investigations on SARS-CoV-1 and MERS-CoV, the approved vaccines from biontech/Pfizer, Moderna, Oxford/Astrazeneca and others under development against SARSCoV- 2 infection. Curr Med Chem 29:4–18. doi: 10.2174/0929867328666210521164809 - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

