Coordination of RAB-8 and RAB-11 during unconventional protein secretion
- PMID: 38019180
- PMCID: PMC10686230
- DOI: 10.1083/jcb.202306107
Coordination of RAB-8 and RAB-11 during unconventional protein secretion
Abstract
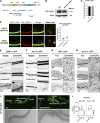
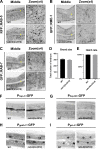
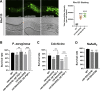
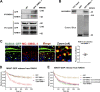
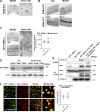
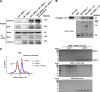
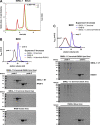
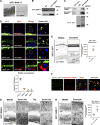
Multiple physiology-pertinent transmembrane proteins reach the cell surface via the Golgi-bypassing unconventional protein secretion (UcPS) pathway. By employing C. elegans-polarized intestine epithelia, we recently have revealed that the small GTPase RAB-8/Rab8 serves as an important player in the process. Nonetheless, its function and the relevant UcPS itinerary remain poorly understood. Here, we show that deregulated RAB-8 activity resulted in impaired apical UcPS, which increased sensitivity to infection and environmental stress. We also identified the SNARE VTI-1/Vti1a/b as a new RAB-8-interacting factor involved in the apical UcPS. Besides, RAB-11/Rab11 was capable of recruiting RABI-8/Rabin8 to reduce the guanine nucleotide exchange activity of SMGL-1/GEF toward RAB-8, indicating the necessity of a finely tuned RAB-8/RAB-11 network. Populations of RAB-8- and RAB-11-positive endosomal structures containing the apical UcPS cargo moved toward the apical side. In the absence of RAB-11 or its effectors, the cargo was retained in RAB-8- and RAB-11-positive endosomes, respectively, suggesting that these endosomes are utilized as intermediate carriers for the UcPS.
© 2023 Li et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures


Comment in
-
A RAB transition orchestrates membrane trafficking in unconventional protein secretion.J Cell Biol. 2024 Feb 5;223(2):e202312096. doi: 10.1083/jcb.202312096. Epub 2024 Jan 5. J Cell Biol. 2024. PMID: 38180797 Free PMC article.
Similar articles
-
RAB-10 cooperates with EHBP-1 to capture vesicular carriers during post-Golgi exocytic trafficking.J Cell Biol. 2025 Apr 7;224(4):e202410003. doi: 10.1083/jcb.202410003. Epub 2025 Feb 21. J Cell Biol. 2025. PMID: 39982707
-
SMGL-1/NBAS acts as a RAB-8 GEF to regulate unconventional protein secretion.J Cell Biol. 2022 Jul 4;221(7):e202111125. doi: 10.1083/jcb.202111125. Epub 2022 May 23. J Cell Biol. 2022. PMID: 35604368 Free PMC article.
-
RAB-10-GTPase-mediated regulation of endosomal phosphatidylinositol-4,5-bisphosphate.Proc Natl Acad Sci U S A. 2012 Aug 28;109(35):E2306-15. doi: 10.1073/pnas.1205278109. Epub 2012 Aug 6. Proc Natl Acad Sci U S A. 2012. PMID: 22869721 Free PMC article.
-
Who's in control? Principles of Rab GTPase activation in endolysosomal membrane trafficking and beyond.J Cell Biol. 2021 Sep 6;220(9):e202105120. doi: 10.1083/jcb.202105120. Epub 2021 Aug 12. J Cell Biol. 2021. PMID: 34383013 Free PMC article. Review.
-
Multiple Roles of VARP in Endosomal Trafficking: Rabs, Retromer Components and R-SNARE VAMP7 Meet on VARP.Traffic. 2016 Jul;17(7):709-19. doi: 10.1111/tra.12406. Epub 2016 May 13. Traffic. 2016. PMID: 27103185 Review.
Cited by
-
An RNAi screen of Rab GTPase genes in Caenorhabditis elegans reveals that morphogenesis has a higher demand than stem cell niche maintenance for rab-1 in the somatic cells of the reproductive system.G3 (Bethesda). 2025 Jun 4;15(6):jkaf085. doi: 10.1093/g3journal/jkaf085. G3 (Bethesda). 2025. PMID: 40239019 Free PMC article.
-
An RNAi screen of Rab GTPase genes in C. elegans reveals that somatic cells of the reproductive system depend on rab-1 for morphogenesis but not stem cell niche maintenance.bioRxiv [Preprint]. 2024 Dec 6:2024.12.03.626641. doi: 10.1101/2024.12.03.626641. bioRxiv. 2024. Update in: G3 (Bethesda). 2025 Jun 4;15(6):jkaf085. doi: 10.1093/g3journal/jkaf085. PMID: 39677816 Free PMC article. Updated. Preprint.
-
RAB-10 cooperates with EHBP-1 to capture vesicular carriers during post-Golgi exocytic trafficking.J Cell Biol. 2025 Apr 7;224(4):e202410003. doi: 10.1083/jcb.202410003. Epub 2025 Feb 21. J Cell Biol. 2025. PMID: 39982707
-
Molecular mechanisms of polarized transport to the apical plasma membrane.Front Cell Dev Biol. 2024 Sep 26;12:1477173. doi: 10.3389/fcell.2024.1477173. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39445332 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

