The putatively high-altitude adaptation of macaque monkeys: Evidence from the fecal metabolome and gut microbiome
- PMID: 38020871
- PMCID: PMC10660799
- DOI: 10.1111/eva.13595
The putatively high-altitude adaptation of macaque monkeys: Evidence from the fecal metabolome and gut microbiome
Abstract
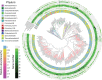
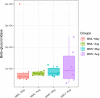
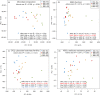
Animals living in high-altitude environments, such as the Tibetan Plateau, must face harsh environmental conditions (e.g., hypoxia, cold, and strong UV radiation). These animals' physiological adaptations (e.g., increased red cell production and turnover rate) might also be associated with the gut microbial response. Bilirubin is a component of red blood cell turnover or destruction and is excreted into the intestine and reduced to urobilinoids and/or urobilinogen by gut bacteria. Here, we found that the feces of macaques living in high-altitude regions look significantly browner (with a high concentration of stercobilin, a component from urobilinoids) than those living in low-altitude regions. We also found that gut microbes involved in urobilinogen reduction (e.g., beta-glucuronidase) were enriched in the high-altitude mammal population compared to the low-altitude population. Moreover, the spatial-temporal change in gut microbial function was more profound in the low-altitude macaques than in the high-altitude population, which might be attributed to profound changes in food resources in the low-altitude regions. Therefore, we conclude that a high-altitude environment's stress influences living animals and their symbiotic microbiota.
Keywords: adaptation; fecal color; gut microbiome; high altitude; primates; spatial–temporal changes.
© 2023 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Amato, K. R. , Leigh, S. R. , Kent, A. , Mackie, R. I. , Yeoman, C. J. , Stumpf, R. M. , Wilson, B. A. , Nelson, K. E. , White, B. A. , & Garber, P. A. (2015). The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microbial Ecology, 69, 434–443. - PubMed
-
- Anderson, M. J. (2001). A new method for non‐parametric multivariate analysis of variance. Austral Ecology, 26, 32–46.
-
- Askew, E. (2002). Work at high altitude and oxidative stress: Antioxidant nutrients. Toxicology, 180, 107–119. - PubMed
LinkOut - more resources
Full Text Sources

