Integrated analysis of single-cell RNA-seq and bulk RNA-seq reveals RNA N6-methyladenosine modification associated with prognosis and drug resistance in acute myeloid leukemia
- PMID: 38022588
- PMCID: PMC10644381
- DOI: 10.3389/fimmu.2023.1281687
Integrated analysis of single-cell RNA-seq and bulk RNA-seq reveals RNA N6-methyladenosine modification associated with prognosis and drug resistance in acute myeloid leukemia
Abstract
Introduction: Acute myeloid leukemia (AML) is a type of blood cancer that is identified by the unrestricted growth of immature myeloid cells within the bone marrow. Despite therapeutic advances, AML prognosis remains highly variable, and there is a lack of biomarkers for customizing treatment. RNA N6-methyladenosine (m6A) modification is a reversible and dynamic process that plays a critical role in cancer progression and drug resistance.
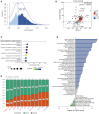
Methods: To investigate the m6A modification patterns in AML and their potential clinical significance, we used the AUCell method to describe the m6A modification activity of cells in AML patients based on 23 m6A modification enzymes and further integrated with bulk RNA-seq data.
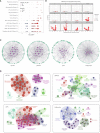
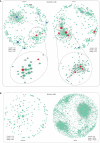
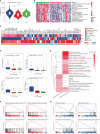
Results: We found that m6A modification was more effective in leukemic cells than in immune cells and induced significant changes in gene expression in leukemic cells rather than immune cells. Furthermore, network analysis revealed a correlation between transcription factor activation and the m6A modification status in leukemia cells, while active m6A-modified immune cells exhibited a higher interaction density in their gene regulatory networks. Hierarchical clustering based on m6A-related genes identified three distinct AML subtypes. The immune dysregulation subtype, characterized by RUNX1 mutation and KMT2A copy number variation, was associated with a worse prognosis and exhibited a specific gene expression pattern with high expression level of IGF2BP3 and FMR1, and low expression level of ELAVL1 and YTHDF2. Notably, patients with the immune dysregulation subtype were sensitive to immunotherapy and chemotherapy.
Discussion: Collectively, our findings suggest that m6A modification could be a potential therapeutic target for AML, and the identified subtypes could guide personalized therapy.
Keywords: N6-methyladenosine modification; acute myeloid leukemia; single-cell transcriptome; subtype; tumor microenvironment.
Copyright © 2023 Li, Liu, Wang, Zhao, Wang, Yu, Wu, Chu and Han.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Comment on
-
Immune dysfunction signatures predict outcomes and define checkpoint blockade-unresponsive microenvironments in acute myeloid leukemia.J Clin Invest. 2022 Nov 1;132(21):e159579. doi: 10.1172/JCI159579. J Clin Invest. 2022. PMID: 36099049 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

