TGFβ-induced circLTBP2 predicts a poor prognosis in intrahepatic cholangiocarcinoma and mediates gemcitabine resistance by sponging miR-338-3p
- PMID: 38023605
- PMCID: PMC10665948
- DOI: 10.1016/j.jhepr.2023.100900
TGFβ-induced circLTBP2 predicts a poor prognosis in intrahepatic cholangiocarcinoma and mediates gemcitabine resistance by sponging miR-338-3p
Abstract
Background & aims: Intrahepatic cholangiocarcinoma (iCCA) is a deadly cancer worldwide with an increasing incidence and limited therapeutic options. Therefore, there is an urgent need to open the field to new concepts for identifying clinically relevant therapeutic targets and biomarkers. Here, we explored the role and the clinical relevance of circular RNA (circRNA) circLTBP2 in iCCA.
Methods: Transforming growth factor β (TGFβ)-regulated circRNAs were identified by dedicated microarrays in human HuCC-T1 iCCA cell line, and their clinical relevance was evaluated in independent cohorts of patients. Gain and loss of function of circLTBP2 combined with functional tests was performed in vitro and in vivo in mice. RNA pulldown, microRNA sequencing, and RNA immunoprecipitation were performed to explore the sponging activity of circLTBP2.
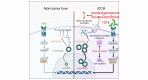
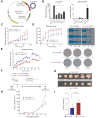
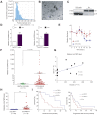
Results: CircLTBP2 (has_circ_0032603) was identified as a novel TGFβ-induced circRNA in several cholangiocarcinoma cell lines. CircLTBP2 promotes tumour cell proliferation, migration, and resistance to gemcitabine-induced apoptosis in vitro and tumour growth in vivo. Mechanistically, circLTBP2 acts as a competitive RNA regulating notably the activity of the tumour suppressor microRNA miR-338-3p, leading to the overexpression of its pro-metastatic targets. The restoration of miR-338-3p levels in iCCA cells reversed the pro-tumourigenic effects driven by circLTBP2, including the resistance to gemcitabine-induced apoptosis. In addition, circLTBP2 expression predicted a reduced survival, as detected in not only tumour tissues but also serum extracellular vesicles isolated from patients with iCCA.
Conclusions: CircLTBP2 is a novel effector of the pro-tumourigenic arm of TGFβ and a clinically relevant biomarker easily detected from liquid biopsies in iCCA.
Impact and implications: Intrahepatic cholangiocarcinoma (iCCA) is an aggressive cancer with limited therapeutic options. Opening the field to new concepts is urgently needed to improve the survival of patients. Here, we evaluated the role and the clinical relevance of circular RNA. We report that TGFβ-induced circLTBP2 contributes to CCA carcinogenesis and may constitute a clinically relevant prognostic biomarker detected in liquid biopsies.
Keywords: Biomarker; Cholangiocarcinoma; Circular RNA; Extracellular vesicles; Transforming growth factor beta.
© 2023 The Authors.
Conflict of interest statement
JE received honoraria from MSD, Roche, AstraZeneca, and BMS. The authors have no other relevant affiliations or financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership, or options, expert testimony, grants or patents received or pending, or royalties. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures



References
-
- Valle J.W., Kelley R.K., Nervi B., Oh D.Y., Zhu A.X. Biliary tract cancer. Lancet. 2021;397:428–444. - PubMed
-
- Izquierdo-Sanchez L., Lamarca A., La Casta A., Buettner S., Utpatel K., Klüumpen H.J., et al. Cholangiocarcinoma landscape in Europe: diagnostic, prognostic and therapeutic insights from the ENSCCA Registry. J Hepatol. 2022;76:1109–1121. - PubMed
-
- Macias R.I.R., Cardinale V., Kendall T.J., Avila M.A., Guido M., Coulouarn C., et al. Clinical relevance of biomarkers in cholangiocarcinoma: critical revision and future directions. Gut. 2022;71:1669–1683. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

