Transcriptional reprogramming of nucleotide metabolism in response to altered pyrimidine availability in Arabidopsis seedlings
- PMID: 38023851
- PMCID: PMC10652772
- DOI: 10.3389/fpls.2023.1273235
Transcriptional reprogramming of nucleotide metabolism in response to altered pyrimidine availability in Arabidopsis seedlings
Abstract
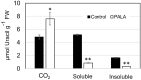
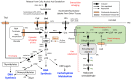
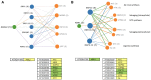
In Arabidopsis seedlings, inhibition of aspartate transcarbamoylase (ATC) and de novo pyrimidine synthesis resulted in pyrimidine starvation and developmental arrest a few days after germination. Synthesis of pyrimidine nucleotides by salvaging of exogenous uridine (Urd) restored normal seedling growth and development. We used this experimental system and transcriptional profiling to investigate genome-wide responses to changes in pyrimidine availability. Gene expression changes at different times after Urd supplementation of pyrimidine-starved seedlings were mapped to major pathways of nucleotide metabolism, in order to better understand potential coordination of pathway activities, at the level of transcription. Repression of de novo synthesis genes and induction of intracellular and extracellular salvaging genes were early and sustained responses to pyrimidine limitation. Since de novo synthesis is energetically more costly than salvaging, this may reflect a reduced energy status of the seedlings, as has been shown in recent studies for seedlings growing under pyrimidine limitation. The unexpected induction of pyrimidine catabolism genes under pyrimidine starvation may result from induction of nucleoside hydrolase NSH1 and repression of genes in the plastid salvaging pathway, diverting uracil (Ura) to catabolism. Identification of pyrimidine-responsive transcription factors with enriched binding sites in highly coexpressed genes of nucleotide metabolism and modeling of potential transcription regulatory networks provided new insights into possible transcriptional control of key enzymes and transporters that regulate nucleotide homeostasis in plants.
Keywords: Arabidopsis; nucleotide metabolism; purine; pyrimidine; transcriptome.
Copyright © 2023 Slocum, Mejia Peña and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures








References
-
- Ashihara H., Ludwig I. A., Crozier A. (2020). Plant nucleotide metabolism ‐ biosynthesis, degradation, and alkaloid formation. doi: 10.1002/9781119476139 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

