Mice with FVB-derived sequence on chromosome 17 succumb to disseminated virus infection due to aberrant NK cell and T cell responses
- PMID: 38026197
- PMCID: PMC10665959
- DOI: 10.1016/j.isci.2023.108348
Mice with FVB-derived sequence on chromosome 17 succumb to disseminated virus infection due to aberrant NK cell and T cell responses
Abstract
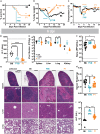
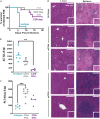
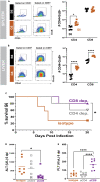
Zoonotic arenavirus infections can result in viral hemorrhagic disease, characterized by platelet loss, petechia, and multi-organ injury. The mechanisms governing these outcomes are likely impacted by virus strain and infection dose, as well as an individual's genetic background and immune constitution. To better understand the processes leading to severe pathogenesis, we compared two strains of inbred mice, C57BL/6J (B6) and FVB/NJ (FVB), that have diametrically opposed outcomes during disseminated lymphocytic choriomeningitis virus (LCMV) infection. Infection caused minimal pathogenesis in B6 mice, whereas FVB mice developed acute hepatitis and perished due, in part, to aberrant NK cell and T cell responses. Susceptible mice showed an outgrowth of cytolytic CD4+ T cells and loss of Treg cells. B6 congenic mice with the FVB allele at a 25Mb locus on chromosome 17 recapitulated FVB pathogenesis upon infection. A locus containing a limited number of variants in immune-related genes greatly impacts survival during infection.
Keywords: Cell biology; Components of the immune system; Genetics; Immunology; Virology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Ogbu O., Ajuluchukwu E., Uneke C.J. Lassa fever in West African sub-region: An overview. J. Vector Borne Dis. 2007;44:1–11. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

