LiDAR Is Effective in Characterizing Vine Growth and Detecting Associated Genetic Loci
- PMID: 38026470
- PMCID: PMC10655830
- DOI: 10.34133/plantphenomics.0116
LiDAR Is Effective in Characterizing Vine Growth and Detecting Associated Genetic Loci
Abstract
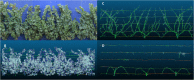
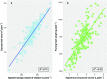
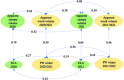
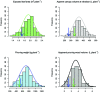
The strong societal demand to reduce pesticide use and adaptation to climate change challenges the capacities of phenotyping new varieties in the vineyard. High-throughput phenotyping is a way to obtain meaningful and reliable information on hundreds of genotypes in a limited period. We evaluated traits related to growth in 209 genotypes from an interspecific grapevine biparental cross, between IJ119, a local genitor, and Divona, both in summer and in winter, using several methods: fresh pruning wood weight, exposed leaf area calculated from digital images, leaf chlorophyll concentration, and LiDAR-derived apparent volumes. Using high-density genetic information obtained by the genotyping by sequencing technology (GBS), we detected 6 regions of the grapevine genome [quantitative trait loci (QTL)] associated with the variations of the traits in the progeny. The detection of statistically significant QTLs, as well as correlations (R2) with traditional methods above 0.46, shows that LiDAR technology is effective in characterizing the growth features of the grapevine. Heritabilities calculated with LiDAR-derived total canopy and pruning wood volumes were high, above 0.66, and stable between growing seasons. These variables provided genetic models explaining up to 47% of the phenotypic variance, which were better than models obtained with the exposed leaf area estimated from images and the destructive pruning weight measurements. Our results highlight the relevance of LiDAR-derived traits for characterizing genetically induced differences in grapevine growth and open new perspectives for high-throughput phenotyping of grapevines in the vineyard.
Copyright © 2023 Elsa Chedid et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures



References
-
- Kraus C, Pennington T, Herzog K, Fisher M, Voegele RT. Effects of canopy architecture and microclimate on grapevine health in two training systems. Vitis. 2018;57(2):53–60.
-
- Valdés-Gómez H, Gary C, Cartolaro P, Lolas-Caneo M, Calonnec A. Powdery mildew development is positively influenced by grapevine vegetative growth induced by different soil management strategies. Cop protection. 2011;30(9):1168–1177.
LinkOut - more resources
Full Text Sources

