Loop-Mediated Isothermal Amplification-Integrated CRISPR Methods for Infectious Disease Diagnosis at Point of Care
- PMID: 38027359
- PMCID: PMC10666231
- DOI: 10.1021/acsomega.3c04422
Loop-Mediated Isothermal Amplification-Integrated CRISPR Methods for Infectious Disease Diagnosis at Point of Care
Abstract
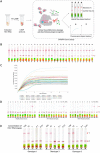
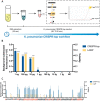
Infectious diseases continue to pose an imminent threat to global public health, leading to high numbers of deaths every year and disproportionately impacting developing countries where access to healthcare is limited. Biological, environmental, and social phenomena, including climate change, globalization, increased population density, and social inequity, contribute to the emergence of novel communicable diseases. Rapid and accurate diagnoses of infectious diseases are essential to preventing the transmission of infectious diseases. Although some commonly used diagnostic technologies provide highly sensitive and specific measurements, limitations including the requirement for complex equipment/infrastructure and refrigeration, the need for trained personnel, long sample processing times, and high cost remain unresolved. To ensure global access to affordable diagnostic methods, loop-mediated isothermal amplification (LAMP) integrated clustered regularly interspaced short palindromic repeat (CRISPR) based pathogen detection has emerged as a promising technology. Here, LAMP-integrated CRISPR-based nucleic acid detection methods are discussed in point-of-care (PoC) pathogen detection platforms, and current limitations and future directions are also identified.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
CRISPR-Cas-Integrated LAMP.Biosensors (Basel). 2022 Nov 17;12(11):1035. doi: 10.3390/bios12111035. Biosensors (Basel). 2022. PMID: 36421156 Free PMC article. Review.
-
Cas12a/Guide RNA-Based Platforms for Rapidly and Accurately Identifying Staphylococcus aureus and Methicillin-Resistant S. aureus.Microbiol Spectr. 2023 Mar 21;11(2):e0487022. doi: 10.1128/spectrum.04870-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36943040 Free PMC article.
-
Clustered Regularly Interspaced short palindromic repeats-Based Microfluidic System in Infectious Diseases Diagnosis: Current Status, Challenges, and Perspectives.Adv Sci (Weinh). 2022 Dec;9(34):e2204172. doi: 10.1002/advs.202204172. Epub 2022 Oct 18. Adv Sci (Weinh). 2022. PMID: 36257813 Free PMC article. Review.
-
SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases.J Clin Microbiol. 2021 Feb 18;59(3):e00745-20. doi: 10.1128/JCM.00745-20. Print 2021 Feb 18. J Clin Microbiol. 2021. Retraction in: J Clin Microbiol. 2024 Jan 17;62(1):e0152123. doi: 10.1128/jcm.01521-23. PMID: 33148705 Free PMC article. Retracted. Review.
-
Integrating CRISPR/Cas within isothermal amplification for point-of-Care Assay of nucleic acid.Talanta. 2022 Jun 1;243:123388. doi: 10.1016/j.talanta.2022.123388. Epub 2022 Mar 12. Talanta. 2022. PMID: 35303554 Review.
Cited by
-
Development and Validation of LAMP Assays for Distinguishing MPXV Clades with Fluorescent and Colorimetric Readouts.Biosensors (Basel). 2025 Jan 6;15(1):23. doi: 10.3390/bios15010023. Biosensors (Basel). 2025. PMID: 39852076 Free PMC article.
-
Revolutionizing Tuberculosis Management With Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas Technology: A Comprehensive Literature Review.Cureus. 2024 Oct 17;16(10):e71697. doi: 10.7759/cureus.71697. eCollection 2024 Oct. Cureus. 2024. PMID: 39552996 Free PMC article. Review.
-
Split crRNA with CRISPR-Cas12a enabling highly sensitive and multiplexed detection of RNA and DNA.Nat Commun. 2024 Sep 27;15(1):8342. doi: 10.1038/s41467-024-52691-x. Nat Commun. 2024. PMID: 39333528 Free PMC article.
-
Harnessing the power of clustered regularly interspaced short palindromic repeats (CRISPR) based microfluidics for next-generation molecular diagnostics.Mol Biol Rep. 2024 Aug 8;51(1):896. doi: 10.1007/s11033-024-09840-8. Mol Biol Rep. 2024. PMID: 39115550 Review.
-
Mpox disease, diagnosis, and point of care platforms.Bioeng Transl Med. 2025 Jan 2;10(3):e10733. doi: 10.1002/btm2.10733. eCollection 2025 May. Bioeng Transl Med. 2025. PMID: 40385539 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

