Evaluation of next generation sequencing approaches for SARS-CoV-2
- PMID: 38027571
- PMCID: PMC10643093
- DOI: 10.1016/j.heliyon.2023.e21101
Evaluation of next generation sequencing approaches for SARS-CoV-2
Abstract
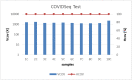
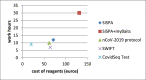
Within public health control strategies for SARS-CoV-2, whole genome sequencing (WGS) is essential for tracking viral spread and monitoring the emergence of variants which may impair the effectiveness of vaccines, diagnostic methods, and therapeutics. In this manuscript different strategies for SARS-CoV-2 WGS including metagenomic shotgun (SG), library enrichment by myBaits® Expert Virus-SARS-CoV-2 (Arbor Biosciences), nCoV-2019 sequencing protocol, ampliseq approach by Swift Amplicon® SARS-CoV-2 Panel kit (Swift Biosciences), and Illumina COVIDSeq Test (Illumina Inc.), were evaluated in order to identify the best approach in terms of results, labour, and costs. The analysis revealed that Illumina COVIDSeq Test (Illumina Inc.) is the best choice for a cost-effective, time-consuming production of consensus sequences.
Keywords: Comparison; Cost; Labor time; Methods; SARS-CoV-2; Whole genome sequencing.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

