Genome-wide identification of glutamate receptor-like gene family in soybean
- PMID: 38027661
- PMCID: PMC10651524
- DOI: 10.1016/j.heliyon.2023.e21655
Genome-wide identification of glutamate receptor-like gene family in soybean
Abstract
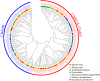
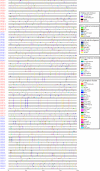
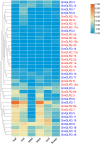
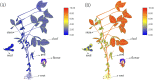
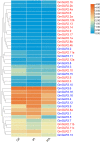
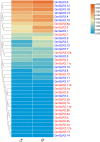
Glutamate receptor-like genes (GLRs) are essential in the growth and development of plants and many physiological and biochemical processes; however, related information in soybean is lacking. In this study, 105 GLRs, including 67 Glycine soja and 38 Glycine max GLRs, were identified and divided into two clades (Clades II and III) according to their phylogenetic relationships. GLR members in the same branch had a relatively conservative motif composition and genetic structure. Furthermore, the soybean GLR family mainly experienced purification selection during evolution. Cis-acting element analysis, gene ontology, and Kyoto Encyclopedia of Genes and Genomic annotations indicated the complexity of the gene regulation and functional diversity of the soybean GLR. Moreover, transcriptome data analysis showed that these GLRs had different expression profiles in different tissues, and Clade III members had higher and more common expression patterns. Additionally, the expression profiles under jasmonic acid treatment and salt stress indicate that the GLR participated in the jasmonic acid signaling pathway and plays a role in salt treatment. This study provides information for a comprehensive understanding of the soybean GLR family and a reference for further functional research and genetic improvement.
Keywords: Expression analysis; Feature analysis; Glutamate receptor-like gene; Soybean.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
LinkOut - more resources
Full Text Sources

