Combined genomic-proteomic approach in the identification of Campylobacter coli amoxicillin-clavulanic acid resistance mechanism in clinical isolates
- PMID: 38029165
- PMCID: PMC10666280
- DOI: 10.3389/fmicb.2023.1285236
Combined genomic-proteomic approach in the identification of Campylobacter coli amoxicillin-clavulanic acid resistance mechanism in clinical isolates
Abstract
Introduction: Aminopenicillins resistance among Campylobacter jejuni and Campylobacter coli strains is associated with a single mutation in the promoting region of a chromosomal beta-lactamase blaOXA61, allowing its expression. Clavulanic acid is used to restore aminopenicillins activity in case of blaOXA61 expression and has also an inherent antimicrobial activity over Campylobacter spp. Resistance to amoxicillin-clavulanic acid is therefore extremely rare among these species: only 0.1% of all Campylobacter spp. analyzed in the French National Reference Center these last years (2017-2022).
Material and methods: Whole genome sequencing with bioinformatic resistance identification combined with mass spectrometry (MS) was used to identify amoxicillin-acid clavulanic resistance mechanism in Campylobacters.
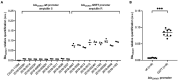
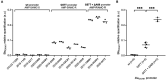
Results: A G57T mutation in blaOXA61 promoting region was identified in all C. jejuni and C. coli ampicillin resistant isolates and no mutation in ampicillin susceptible isolates. Interestingly, three C. coli resistant to both ampicillin and amoxicillin-clavulanic acid displayed a supplemental deletion in the promoting region of blaOXA61 beta-lactamase, at position A69. Using MS, a significant difference in the expression of BlaOXA61 was observed between these three isolates and amoxicillin-clavulanic acid susceptible C. coli.
Conclusion: A combined genomics/proteomics approach allowed here to identify a rare putative resistance mechanism associated with amoxicillin-clavulanic acid resistance for C. coli.
Keywords: AMR; Campylobacter; amoxicillin-clavulanic acid; beta-lactamase; gene expression.
Copyright © 2023 Deforet, Jehanne, Bénéjat, Aptel, Prat, Desbiolles, Ducournau, Jauvain, Bonnet, Vandenesch, Lemoine and Lehours.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Anampa D., Benites C., Lázaro C., Espinoza J., Angulo P., Díaz D., et al. (2020). Detection of the ermB gene associated with macrolide resistance in Campylobacter strains isolated from chickens marketed in Lima, Peru. Rev. Panam. Salud Publica Pan Am. J. Public Health 44:e60. doi: 10.26633/rpsp.2020.60 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

