Unveiling the inhibition mechanism of Clostridioides difficile by Bifidobacterium longum via multiomics approach
- PMID: 38029200
- PMCID: PMC10663266
- DOI: 10.3389/fmicb.2023.1293149
Unveiling the inhibition mechanism of Clostridioides difficile by Bifidobacterium longum via multiomics approach
Abstract
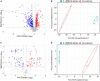
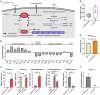
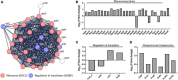
Antibiotic-induced gut microbiota disruption constitutes a major risk factor for Clostridioides difficile infection (CDI). Further, antibiotic therapy, which is the standard treatment option for CDI, exacerbates gut microbiota imbalance, thereby causing high recurrent CDI incidence. Consequently, probiotic-based CDI treatment has emerged as a long-term management and preventive option. However, the mechanisms underlying the therapeutic effects of probiotics for CDI remain uninvestigated, thereby creating a knowledge gap that needs to be addressed. To fill this gap, we used a multiomics approach to holistically investigate the mechanisms underlying the therapeutic effects of probiotics for CDI at a molecular level. We first screened Bifidobacterium longum owing to its inhibitory effect on C. difficile growth, then observed the physiological changes associated with the inhibition of C. difficile growth and toxin production via a multiomics approach. Regarding the mechanism underlying C. difficile growth inhibition, we detected a decrease in intracellular adenosine triphosphate (ATP) synthesis due to B. longum-produced lactate and a subsequent decrease in (deoxy)ribonucleoside triphosphate synthesis. Via the differential regulation of proteins involved in translation and protein quality control, we identified B. longum-induced proteinaceous stress. Finally, we found that B. longum suppressed the toxin production of C. difficile by replenishing proline consumed by it. Overall, the findings of the present study expand our understanding of the mechanisms by which probiotics inhibit C. difficile growth and contribute to the development of live biotherapeutic products based on molecular mechanisms for treating CDI.
Keywords: Bifidobacterium longum; Clostridioides difficile; microbe-microbe interaction; molecular mechanism; multiomics.
Copyright © 2023 Jo, Jeon, Song, Lee, Kwon, Park, Kim, Kim, Baek, Kwon, Kim, Yang and Kim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

