Targeted degrader technologies as prospective SARS-CoV-2 therapies
- PMID: 38029836
- PMCID: PMC10836335
- DOI: 10.1016/j.drudis.2023.103847
Targeted degrader technologies as prospective SARS-CoV-2 therapies
Abstract
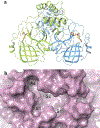
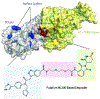
COVID-19 remains a severe public health threat despite the WHO declaring an end to the public health emergency in May 2023. Continual development of SARS-CoV-2 variants with resistance to vaccine-induced or natural immunity necessitates constant vigilance as well as new vaccines and therapeutics. Targeted protein degradation (TPD) remains relatively untapped in antiviral drug discovery and holds the promise of attenuating viral resistance development. From a unique structural design perspective, this review covers antiviral degrader merits and challenges by highlighting key coronavirus protein targets and their co-crystal structures, specifically illustrating how TPD strategies can refine existing SARS-CoV-2 3CL protease inhibitors to potentially produce superior protease-degrading agents.
Keywords: PROTAC; SARS-CoV-2; antiviral; protease; targeted protein degradation.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflicts
The authors have no conflicts to declare.
Declarations of interest
The authors have no conflicts of interest to declare.
Figures



References
-
- Looi MK. Covid-19: scientists sound alarm over new BA.2.86 “Pirola” variant. Br Med J. 2023;382:1964. - PubMed
-
- Abbasi J. What to know about EG.5, the latest SARS-CoV-2 “Variant of Interest”. J Am Med Assoc. 2023;330(10):900–901. - PubMed
-
- Looi MK. Covid-19: hospital admissions rise in England amid fears of new variant and waning immunity. Br Med J. 2023;382:1833. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

