Characterization of the cystargolide biosynthetic gene cluster and functional analysis of the methyltransferase CysG
- PMID: 38029966
- PMCID: PMC10776993
- DOI: 10.1016/j.jbc.2023.105507
Characterization of the cystargolide biosynthetic gene cluster and functional analysis of the methyltransferase CysG
Abstract
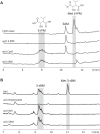
Cystargolides are natural products originally isolated from Kitasatospora cystarginea NRRL B16505 as inhibitors of the proteasome. They are composed of a dipeptide backbone linked to a β-lactone warhead. Recently, we identified the cystargolide biosynthetic gene cluster, but systematic genetic analyses had not been carried out because of the lack of a heterologous expression system. Here, we report the discovery of a homologous cystargolide biosynthetic pathway in Streptomyces durhamensis NRRL-B3309 by genome mining. The gene cluster was cloned via transformation-associated recombination and heterologously expressed in Streptomyces coelicolor M512. We demonstrate that it contains all genes necessary for the production of cystargolide A and B. Single gene deletion experiments reveal that only five of the eight genes from the initially proposed gene cluster are essential for cystargolide synthesis. Additional insights into the cystargolide pathway could be obtained from in vitro assays with CysG and chemical complementation of the respective gene knockout. This could be further supported by the in vitro investigation of the CysG homolog BelI from the belactosin biosynthetic gene cluster. Thereby, we confirm that CysG and BelI catalyze a cryptic SAM-dependent transfer of a methyl group that is critical for the construction of the cystargolide and belactosin β-lactone warheads.
Keywords: biosynthesis; gene knockout; heterologous expression; inhibitor; lactone; natural product; proteasome.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no conflicts of interest with the contents of the article.
Figures
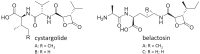


References
-
- Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H.D., et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. - PubMed
-
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. - PubMed
-
- Clurman B.E., Sheaff R.J., Thress K., Groudine M., Roberts J.M. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes. Dev. 1996;10:1979–1990. - PubMed
-
- Orlowski R.Z. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 1999;6:303–313. - PubMed
-
- Rock K.L., Goldberg A.L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 1999;17:739–779. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

