TRS: a method for determining transcript termini from RNAtag-seq sequencing data
- PMID: 38030608
- PMCID: PMC10687069
- DOI: 10.1038/s41467-023-43534-2
TRS: a method for determining transcript termini from RNAtag-seq sequencing data
Abstract
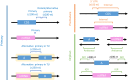
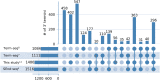
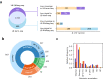
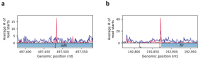
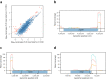
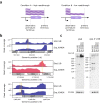
In bacteria, determination of the 3' termini of transcripts plays an essential role in regulation of gene expression, affecting the functionality and stability of the transcript. Several experimental approaches were developed to identify the 3' termini of transcripts, however, these were applied only to a limited number of bacteria and growth conditions. Here we present a straightforward approach to identify 3' termini from widely available RNA-seq data without the need for additional experiments. Our approach relies on the observation that the RNAtag-seq sequencing protocol results in overabundance of reads mapped to transcript 3' termini. We present TRS (Termini by Read Starts), a computational pipeline exploiting this property to identify 3' termini in RNAtag-seq data, and show that the identified 3' termini are highly reliable. Since RNAtag-seq data are widely available for many bacteria and growth conditions, our approach paves the way for studying bacterial transcription termination in an unprecedented scope.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

