Contribution of pks+ E. coli mutations to colorectal carcinogenesis
- PMID: 38030613
- PMCID: PMC10687070
- DOI: 10.1038/s41467-023-43329-5
Contribution of pks+ E. coli mutations to colorectal carcinogenesis
Abstract
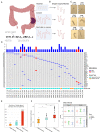
The dominant mutational signature in colorectal cancer genomes is C > T deamination (COSMIC Signature 1) and, in a small subgroup, mismatch repair signature (COSMIC signatures 6 and 44). Mutations in common colorectal cancer driver genes are often not consistent with those signatures. Here we perform whole-genome sequencing of normal colon crypts from cancer patients, matched to a previous multi-omic tumour dataset. We analyse normal crypts that were distant vs adjacent to the cancer. In contrast to healthy individuals, normal crypts of colon cancer patients have a high incidence of pks + (polyketide synthases) E.coli (Escherichia coli) mutational and indel signatures, and this is confirmed by metagenomics. These signatures are compatible with many clonal driver mutations detected in the corresponding cancer samples, including in chromatin modifier genes, supporting their role in early tumourigenesis. These results provide evidence that pks + E.coli is a potential driver of carcinogenesis in the human gut.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

