Gene interaction network analysis in multiple myeloma detects complex immune dysregulation associated with shorter survival
- PMID: 38030619
- PMCID: PMC10687027
- DOI: 10.1038/s41408-023-00935-2
Gene interaction network analysis in multiple myeloma detects complex immune dysregulation associated with shorter survival
Abstract
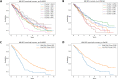
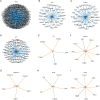
The plasma cell cancer multiple myeloma (MM) varies significantly in genomic characteristics, response to therapy, and long-term prognosis. To investigate global interactions in MM, we combined a known protein interaction network with a large clinically annotated MM dataset. We hypothesized that an unbiased network analysis method based on large-scale similarities in gene expression, copy number aberration, and protein interactions may provide novel biological insights. Applying a novel measure of network robustness, Ollivier-Ricci Curvature, we examined patterns in the RNA-Seq gene expression and CNA data and how they relate to clinical outcomes. Hierarchical clustering using ORC differentiated high-risk subtypes with low progression free survival. Differential gene expression analysis defined 118 genes with significantly aberrant expression. These genes, while not previously associated with MM, were associated with DNA repair, apoptosis, and the immune system. Univariate analysis identified 8/118 to be prognostic genes; all associated with the immune system. A network topology analysis identified both hub and bridge genes which connect known genes of biological significance of MM. Taken together, gene interaction network analysis in MM uses a novel method of global assessment to demonstrate complex immune dysregulation associated with shorter survival.
© 2023. The Author(s).
Conflict of interest statement
SZU: Research funding: Amgen, BMS/Celgene, GSK, Janssen, Merck, Pharmacyclics, Sanofi, Seattle Genetics, Takeda. Consulting/Advisory Board: Abbvie, Amgen, BMS, Celgene, Genentech, Gilead, GSK, Janssen, Sanofi, Seattle Genetics, SecuraBio, SkylineDX, Takeda, TeneoBio.
Figures




References
-
- D’Agostino M, Cairns DA, Lahuerta JJ, Wester R, Bertsch U, Waage A, et al. Second revision of the International Staging System (R2-ISS) for overall survival in multiple myeloma: a European Myeloma Network (EMN) report within the HARMONY project. J Clin Oncol. 2022;40:3406–18. doi: 10.1200/JCO.21.02614. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

