Immunosignatures associated with TP53 status and co-mutations classify prognostically head and neck cancer patients
- PMID: 38031025
- PMCID: PMC10687972
- DOI: 10.1186/s12943-023-01905-9
Immunosignatures associated with TP53 status and co-mutations classify prognostically head and neck cancer patients
Abstract
Background: Immune checkpoint inhibitors (ICIs) are a therapeutic strategy for various cancers although only a subset of patients respond to the therapy. Identifying patients more prone to respond to ICIs may increase the therapeutic benefit and allow studying new approaches for resistant patients.
Methods: We analyzed the TCGA cohort of HNSCC patients in relation to their activation of 26 immune gene expression signatures, as well as their cell type composition, in order to define signaling pathways associated with resistance to ICIs. Results were validated on two cohorts of 102 HNSCC patients and 139 HNSCC patients under treatment with PD-L1 inhibitors, respectively, and a cohort of 108 HNSCC HPV negative patients and by in vitro experiments in HNSCC cell lines.
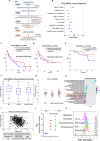
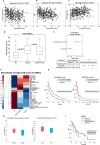
Results: We observed a significant association between the gene set and TP53 gene status and OS and PFS of HNSCC patients. Surprisingly, the presence of a TP53 mutation together with another co-driver mutation was associated with significantly higher levels of the immune gene expression, in comparison to tumors in which the TP53 gene was mutated alone. In addition, the higher level of TP53 mutated-dependent MYC signature was associated with lower levels of the immune gene expression signature. In vitro and three different patient cohorts validation analyses corroborated these findings.
Conclusions: Immune gene signature sets associated with TP53 status and co-mutations classify with more accuracy HNSCC patients. These biomarkers may be easily implemented in clinical setting.
Keywords: HNSCC; Immune checkpoint inhibitor; Immunotherapy; PDL1; PI3K; c-MYC; p53.
© 2023. The Author(s).
Conflict of interest statement
He authors declare no competing interests.
Figures
References
-
- Machiels JP, Rene Leemans C, Golusinski W, Grau C, Licitra L, Gregoire V, secretariat@ehns.org EEBEa, clinicalguidelines@esmo.org EGCEa, info@estro.org EEBEa. Squamous cell carcinoma of the oral cavity, larynx, oropharynx and hypopharynx: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31:1462–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

