Integrating compositional and functional content to describe vaginal microbiomes in health and disease
- PMID: 38031142
- PMCID: PMC10688475
- DOI: 10.1186/s40168-023-01692-x
Integrating compositional and functional content to describe vaginal microbiomes in health and disease
Erratum in
-
Correction: Integrating compositional and functional content to describe vaginal microbiomes in health and disease.Microbiome. 2024 Feb 6;12(1):21. doi: 10.1186/s40168-024-01765-5. Microbiome. 2024. PMID: 38321533 Free PMC article. No abstract available.
Abstract
Background: A Lactobacillus-dominated vaginal microbiome provides the first line of defense against adverse genital tract health outcomes. However, there is limited understanding of the mechanisms by which the vaginal microbiome modulates protection, as prior work mostly described its composition through morphologic assessment and marker gene sequencing methods that do not capture functional information. To address this gap, we developed metagenomic community state types (mgCSTs) which use metagenomic sequences to describe and define vaginal microbiomes based on both composition and functional potential.
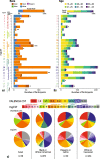
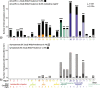
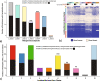
Results: MgCSTs are categories of microbiomes classified using taxonomy and the functional potential encoded in their metagenomes. MgCSTs reflect unique combinations of metagenomic subspecies (mgSs), which are assemblages of bacterial strains of the same species, within a microbiome. We demonstrate that mgCSTs are associated with demographics such as age and race, as well as vaginal pH and Gram stain assessment of vaginal smears. Importantly, these associations varied between mgCSTs predominated by the same bacterial species. A subset of mgCSTs, including three of the six predominated by Gardnerella vaginalis mgSs, as well as mgSs of L. iners, were associated with a greater likelihood of bacterial vaginosis diagnosed by Amsel clinical criteria. This L. iners mgSs, among other functional features, encoded enhanced genetic capabilities for epithelial cell attachment that could facilitate cytotoxin-mediated cell lysis. Finally, we report a mgSs and mgCST classifier for which source code is provided and may be adapted for use by the microbiome research community.
Conclusions: MgCSTs are a novel and easily implemented approach to reduce the dimension of complex metagenomic datasets while maintaining their functional uniqueness. MgCSTs enable the investigation of multiple strains of the same species and the functional diversity in that species. Future investigations of functional diversity may be key to unraveling the pathways by which the vaginal microbiome modulates the protection of the genital tract. Importantly, our findings support the hypothesis that functional differences between vaginal microbiomes, including those that may look compositionally similar, are critical considerations in vaginal health. Ultimately, mgCSTs may lead to novel hypotheses concerning the role of the vaginal microbiome in promoting health and disease, and identify targets for novel prognostic, diagnostic, and therapeutic strategies to improve women's genital health. Video Abstract.
Keywords: Bacterial vaginosis; Genital health; Metagenome; Sequencing; Vaginal microbiome.
© 2023. The Author(s).
Conflict of interest statement
JR is co-founder of LUCA Biologics, a biotechnology company focusing on translating microbiome research into live biotherapeutics drugs for women’s health. JR is Editor-in-Chief at
Figures





Update of
-
High-resolution functional description of vaginal microbiomes in health and disease.bioRxiv [Preprint]. 2023 Mar 24:2023.03.24.533147. doi: 10.1101/2023.03.24.533147. bioRxiv. 2023. Update in: Microbiome. 2023 Nov 30;11(1):259. doi: 10.1186/s40168-023-01692-x. PMID: 36993583 Free PMC article. Updated. Preprint.
References
-
- Witkin SS, Mendes-Soares H, Linhares IM, Jayaram A, Ledger WJ, Forney LJ. Influence of vaginal bacteria and D- and L-lactic acid isomers on vaginal extracellular matrix metalloproteinase inducer: implications for protection against upper genital tract infections. mBio. 2013;4(4); 10.1128/mBio.00460-13. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

