Unveiling the role of osteosarcoma-derived secretome in premetastatic lung remodelling
- PMID: 38031171
- PMCID: PMC10688015
- DOI: 10.1186/s13046-023-02886-9
Unveiling the role of osteosarcoma-derived secretome in premetastatic lung remodelling
Abstract
Background: Lung metastasis is the most adverse clinical factor and remains the leading cause of osteosarcoma-related death. Deciphering the mechanisms driving metastatic spread is crucial for finding open therapeutic windows for successful organ-specific interventions that may halt or prevent lung metastasis.
Methods: We employed a mouse premetastatic lung-based multi-omics integrative approach combined with clinical features to uncover the specific changes that precede lung metastasis formation and identify novel molecular targets and biomarker of clinical utility that enable the design of novel therapeutic strategies.
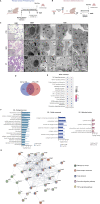
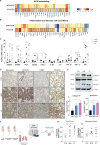
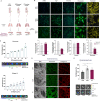
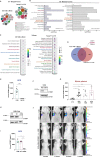
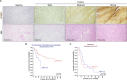
Results: We found that osteosarcoma-bearing mice or those preconditioned with the osteosarcoma cell secretome harbour profound lung structural alterations with airway damage, inflammation, neutrophil infiltration, and extracellular matrix remodelling with increased deposition of fibronectin and collagens by resident stromal activated fibroblasts, favouring the adhesion of disseminated tumour cells. Systemic-induced microenvironmental changes, supported by transcriptomic and histological data, promoted and accelerated lung metastasis formation. Comparative proteome profiling of the cell secretome and mouse plasma identified a large number of proteins involved in extracellular-matrix organization, cell-matrix adhesion, neutrophil degranulation, and cytokine-mediated signalling, consistent with the observed lung microenvironmental changes. Moreover, we identified EFEMP1, an extracellular matrix glycoprotein exclusively secreted by metastatic cells, in the plasma of mice bearing a primary tumour and in biopsy specimens from osteosarcoma patients with poorer overall survival. Depletion of EFEMP1 from the secretome prevents the formation of lung metastasis.
Conclusions: Integration of our data uncovers neutrophil infiltration and the functional contribution of stromal-activated fibroblasts in ECM remodelling for tumour cell attachment as early pro-metastatic events, which may hold therapeutic potential in preventing or slowing the metastatic spread. Moreover, we identified EFEMP1, a secreted glycoprotein, as a metastatic driver and a potential candidate prognostic biomarker for lung metastasis in osteosarcoma patients. Osteosarcoma-derived secreted factors systemically reprogrammed the lung microenvironment and fostered a growth-permissive niche for incoming disseminated cells to survive and outgrow into overt metastasis. Daily administration of osteosarcoma cell secretome mimics the systemic release of tumour-secreted factors of a growing tumour in mice during PMN formation; Transcriptomic and histological analysis of premetastatic lungs revealed inflammatory-induced stromal fibroblast activation, neutrophil infiltration, and ECM remodelling as early onset pro-metastatic events; Proteome profiling identified EFEMP1, an extracellular secreted glycoprotein, as a potential predictive biomarker for lung metastasis and poor prognosis in osteosarcoma patients. Osteosarcoma patients with EFEMP1 expressing biopsies have a poorer overall survival.
Keywords: EFEMP1; Extracellular matrix; Fibroblasts; Lung Metastasis; Neutrophils; Osteosarcoma; Premetastatic niche.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

