Deep phenotyping characterization of human unconventional CD8+NKG2A/C+ T cells among T and NK cells by spectral flow cytometry
- PMID: 38032799
- PMCID: PMC10711235
- DOI: 10.1016/j.xpro.2023.102734
Deep phenotyping characterization of human unconventional CD8+NKG2A/C+ T cells among T and NK cells by spectral flow cytometry
Abstract
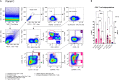
Here, we present a protocol for setting three spectral flow cytometry panels for the characterization of human unconventional CD8+NKG2A/C+ T cells as well as other T and natural killer cell subsets. We describe steps for standardizing, preparing, and staining the cells, the experimental setup, and the final data analysis. This protocol should be advantageous in various settings including immunophenotyping of limited samples, immune function evaluation/monitoring, as well as research in oncology, autoimmune, and infectious diseases.
Keywords: Antibody; Flow Cytometry; Immunology.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Braud V.M., Allan D.S., O’Callaghan C.A., Söderström K., D’Andrea A., Ogg G.S., Lazetic S., Young N.T., Bell J.I., Phillips J.H., et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. - PubMed
-
- Li W.-X., Pan H.-F., Hu J.-L., Wang C.-Z., Zhang N., Li J., Li X.-P., Xu J.-H., Ye D.-Q. Assay of T- and NK-cell subsets and the expression of NKG2A and NKG2D in patients with new-onset systemic lupus erythematosus. Clin. Rheumatol. 2010;29:315–323. - PubMed
-
- Kasakovski D., Zeng X., Lai J., Yu Z., Yao D., Chen S., Zha X., Li Y., Xu L. Characterization of KIR + NKG2A + Eomes- NK-like CD8+ T cells and their decline with age in healthy individuals. Cytometry B Clin. Cytom. 2021;100:467–475. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

