Representing true plant genomes: haplotype-resolved hybrid pepper genome with trio-binning
- PMID: 38034563
- PMCID: PMC10687446
- DOI: 10.3389/fpls.2023.1184112
Representing true plant genomes: haplotype-resolved hybrid pepper genome with trio-binning
Abstract
As sequencing costs decrease and availability of high fidelity long-read sequencing increases, generating experiment specific de novo genome assemblies becomes feasible. In many crop species, obtaining the genome of a hybrid or heterozygous individual is necessary for systems that do not tolerate inbreeding or for investigating important biological questions, such as hybrid vigor. However, most genome assembly methods that have been used in plants result in a merged single sequence representation that is not a true biologically accurate representation of either haplotype within a diploid individual. The resulting genome assembly is often fragmented and exhibits a mosaic of the two haplotypes, referred to as haplotype-switching. Important haplotype level information, such as causal mutations and structural variation is therefore lost causing difficulties in interpreting downstream analyses. To overcome this challenge, we have applied a method developed for animal genome assembly called trio-binning to an intra-specific hybrid of chili pepper (Capsicum annuum L. cv. HDA149 x Capsicum annuum L. cv. HDA330). We tested all currently available softwares for performing trio-binning, combined with multiple scaffolding technologies including Bionano to determine the optimal method of producing the best haplotype-resolved assembly. Ultimately, we produced highly contiguous biologically true haplotype-resolved genome assemblies for each parent, with scaffold N50s of 266.0 Mb and 281.3 Mb, with 99.6% and 99.8% positioned into chromosomes respectively. The assemblies captured 3.10 Gb and 3.12 Gb of the estimated 3.5 Gb chili pepper genome size. These assemblies represent the complete genome structure of the intraspecific hybrid, as well as the two parental genomes, and show measurable improvements over the currently available reference genomes. Our manuscript provides a valuable guide on how to apply trio-binning to other plant genomes.
Keywords: HiFi; genome assembly; haplotype; pepper; trio-binning.
Copyright © 2023 Delorean, Youngblood, Simpson, Schoonmaker, Scheffler, Rutter and Hulse-Kemp.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


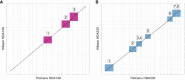
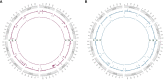
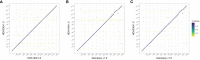
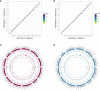
References
-
- Andrews S. (2010) FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
-
- Belletti P., Marzachì C., Lanteri S. (1998). Flow cytometric measurement of nuclear DNA content in Capsicum (Solanaceae). Plant System. Evol. 209, 85–91. doi: 10.1007/BF00991526 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

