MANGEM: A web app for multimodal analysis of neuronal gene expression, electrophysiology, and morphology
- PMID: 38035195
- PMCID: PMC10682747
- DOI: 10.1016/j.patter.2023.100847
MANGEM: A web app for multimodal analysis of neuronal gene expression, electrophysiology, and morphology
Abstract
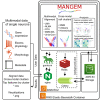
Single-cell techniques like Patch-seq have enabled the acquisition of multimodal data from individual neuronal cells, offering systematic insights into neuronal functions. However, these data can be heterogeneous and noisy. To address this, machine learning methods have been used to align cells from different modalities onto a low-dimensional latent space, revealing multimodal cell clusters. The use of those methods can be challenging without computational expertise or suitable computing infrastructure for computationally expensive methods. To address this, we developed a cloud-based web application, MANGEM (multimodal analysis of neuronal gene expression, electrophysiology, and morphology). MANGEM provides a step-by-step accessible and user-friendly interface to machine learning alignment methods of neuronal multimodal data. It can run asynchronously for large-scale data alignment, provide users with various downstream analyses of aligned cells, and visualize the analytic results. We demonstrated the usage of MANGEM by aligning multimodal data of neuronal cells in the mouse visual cortex.
Keywords: asynchronous computation; cloud-based machine learning; cross-modal cell clusters and phenotypes; gene expression; manifold learning; multimodal data alignment; neuronal electrophysiology and morphology; patch-seq analysis; single-cell multimodalities; web application.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
MANGEM: a web app for Multimodal Analysis of Neuronal Gene expression, Electrophysiology and Morphology.bioRxiv [Preprint]. 2023 Apr 4:2023.04.03.535322. doi: 10.1101/2023.04.03.535322. bioRxiv. 2023. Update in: Patterns (N Y). 2023 Sep 25;4(11):100847. doi: 10.1016/j.patter.2023.100847. PMID: 37066386 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources

