Evaluation of tissue- and plasma-derived tumor mutational burden (TMB) and genomic alterations of interest in CheckMate 848, a study of nivolumab combined with ipilimumab and nivolumab alone in patients with advanced or metastatic solid tumors with high TMB
- PMID: 38035725
- PMCID: PMC10689409
- DOI: 10.1136/jitc-2023-007339
Evaluation of tissue- and plasma-derived tumor mutational burden (TMB) and genomic alterations of interest in CheckMate 848, a study of nivolumab combined with ipilimumab and nivolumab alone in patients with advanced or metastatic solid tumors with high TMB
Abstract
Background: An accumulation of somatic mutations in tumors leads to increased neoantigen levels and antitumor immune response. Tumor mutational burden (TMB) reflects the rate of somatic mutations in the tumor genome, as determined from tumor tissue (tTMB) or blood (bTMB). While high tTMB is a biomarker of immune checkpoint inhibitor (ICI) treatment efficacy, few studies have explored the clinical utility of bTMB, a less invasive alternative for TMB assessment. Establishing the correlation between tTMB and bTMB would provide insight into whether bTMB is a potential substitute for tTMB. We explored the tumor genomes of patients enrolled in CheckMate 848 with measurable TMB. The correlation between tTMB and bTMB, and the factors affecting it, were evaluated.
Methods: In the phase 2 CheckMate 848 (NCT03668119) study, immuno-oncology-naïve patients with advanced, metastatic, or unresectable solid tumors and tTMB-high or bTMB-high (≥10 mut/Mb) were prospectively randomized 2:1 to receive nivolumab plus ipilimumab or nivolumab monotherapy. Tissue and plasma DNA sequencing was performed using the Foundation Medicine FoundationOne CDx and bTMB Clinical Trial Assays, respectively. tTMB was quantified from coding variants, insertions, and deletions, and bTMB from somatic base substitutions. Correlations between tTMB and bTMB were determined across samples and with respect to maximum somatic allele frequency (MSAF). Assay agreement and variant composition were also evaluated.
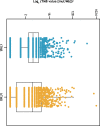
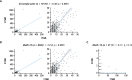
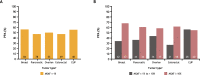
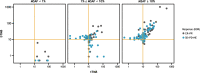
Results: A total of 1,438 and 1,720 unique tissue and blood samples, respectively, were obtained from 1,954 patients and included >100 screened disease ontologies, with 1,017 unique pairs of tTMB and bTMB measurements available for assessment. Median tTMB and bTMB were 3.8 and 3.5 mut/Mb, respectively. A significant correlation between tTMB and bTMB (r=0.48, p<0.0001) was observed across all sample pairs, which increased to r=0.54 (p<0.0001) for samples with MSAF≥1%. Assay concordance was highest for samples with MSAF≥10% across multiple disease ontologies and observed for both responders and non-responders to ICI therapy. The variants contributing to tTMB and bTMB were similar.
Conclusions: We observed that tTMB and bTMB had a statistically significant correlation, particularly for samples with high MSAF, and that this correlation applied across disease ontologies. Further investigation into the clinical utility of bTMB is warranted.
Keywords: Biomarkers, Tumor; Immunotherapy; Ipilimumab; Nivolumab.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: DF, DCP, EME, GRO, HT, JH, and LAA are employees of Foundation Medicine, Inc. and hold stock in Bristol Myers Squibb. GF, GG, and JP are employees of and hold stock in Bristol Myers Squibb. JB is an employee of and holds stock in Bristol Myers Squibb, and holds stock in Johnson and Johnson. NK holds stock in Bristol Myers Squibb. PD is a former employee of Bristol Myers Squibb.
Figures
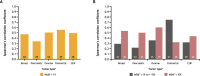
References
-
- Merino DM, McShane LM, Fabrizio D, et al. . Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer 2020;8:e000147. 10.1136/jitc-2019-000147 - DOI - PMC - PubMed
-
- Marabelle A, Fakih M, Lopez J, et al. . Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020;21:1353–65. 10.1016/S1470-2045(20)30445-9 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
