Extracellular Vesicles in Infrapatellar Fat Pad from Osteoarthritis Patients Impair Cartilage Metabolism and Induce Senescence
- PMID: 38036301
- PMCID: PMC10797473
- DOI: 10.1002/advs.202303614
Extracellular Vesicles in Infrapatellar Fat Pad from Osteoarthritis Patients Impair Cartilage Metabolism and Induce Senescence
Abstract
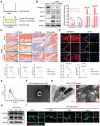
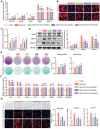
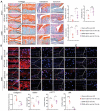
Infrapatellar fat pad (IPFP) is closely associated with the development and progression of knee osteoarthritis (OA), but the underlying mechanism remains unclear. Here, it is find that IPFP from OA patients can secret small extracellular vesicles (sEVs) and deliver them into articular chondrocytes. Inhibition the release of endogenous osteoarthritic IPFP-sEVs by GW4869 significantly alleviated IPFP-sEVs-induced cartilage destruction. Functional assays in vitro demonstrated that IPFP-sEVs significantly promoted chondrocyte extracellular matrix (ECM) catabolism and induced cellular senescence. It is further demonstrated that IPFP-sEVs induced ECM degradation in human and mice cartilage explants and aggravated the progression of experimental OA in mice. Mechanistically, highly enriched let-7b-5p and let-7c-5p in IPFP-sEVs are essential to mediate detrimental effects by directly decreasing senescence negative regulator, lamin B receptor (LBR). Notably, intra-articular injection of antagomirs inhibiting let-7b-5p and let-7c-5p in mice increased LBR expression, suppressed chondrocyte senescence and ameliorated the progression of experimental OA model. This study uncovers the function and mechanism of the IPFP-sEVs in the progression of OA. Targeting IPFP-sEVs cargoes of let-7b-5p and let-7c-5p can provide a potential strategy for OA therapy.
Keywords: extracellular vesicles; infrapatellar fat pad; lamin B receptor; osteoarthritis; senescence.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Hunter D. J., Bierma‐Zeinstra S., Lancet 2019, 393, 1745. - PubMed
-
- Ratneswaran A., Kapoor M., Osteoarthritis cartilage 2021, 29, 151. - PubMed
-
- Quicke J. G., Conaghan P. G., Corp N., Peat G., Osteoarthritis cartilage 2022, 30, 196. - PubMed
-
- Rice S. J., Beier F., Young D. A., Loughlin J., Nat Rev. Rheumatol 2020, 16, 268. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
