A Circular RNA Generated from Nebulin (NEB) Gene Splicing Promotes Skeletal Muscle Myogenesis in Cattle as Detected by a Multi-Omics Approach
- PMID: 38036415
- PMCID: PMC10797441
- DOI: 10.1002/advs.202300702
A Circular RNA Generated from Nebulin (NEB) Gene Splicing Promotes Skeletal Muscle Myogenesis in Cattle as Detected by a Multi-Omics Approach
Abstract
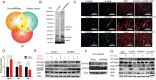
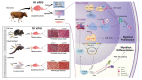
Cattle and the draught force provided by its skeletal muscle have been integral to agro-ecosystems of agricultural civilization for millennia. However, relatively little is known about the cattle muscle functional genomics (including protein coding genes, non-coding RNA, etc.). Circular RNAs (circRNAs), as a new class of non-coding RNAs, can be effectively translated into detectable peptides, which enlightened us on the importance of circRNAs in cattle muscle physiology function. Here, RNA-seq, Ribosome profiling (Ribo-seq), and peptidome data are integrated from cattle skeletal muscle, and detected five encoded peptides from circRNAs. It is further identified and functionally characterize a 907-amino acids muscle-specific peptide that is named circNEB-peptide because derived by the splicing of Nebulin (NEB) gene. This peptide localizes to the nucleus and cytoplasm and directly interacts with SKP1 and TPM1, key factors regulating physiological activities of myoblasts, via ubiquitination and myoblast fusion, respectively. The circNEB-peptide is found to promote myoblasts proliferation and differentiation in vitro, and induce muscle regeneration in vivo. These findings suggest circNEB-peptide is an important regulator of skeletal muscle regeneration and underscore the possibility that more encoding polypeptides derived by RNAs currently annotated as non-coding exist.
Keywords: Cattle; Ribo-seq; SCF complex; circNEB; myogenesis; skeletal muscle.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
