Bile proteome reveals biliary regeneration during normothermic preservation of human donor livers
- PMID: 38036513
- PMCID: PMC10689461
- DOI: 10.1038/s41467-023-43368-y
Bile proteome reveals biliary regeneration during normothermic preservation of human donor livers
Abstract
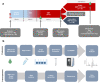
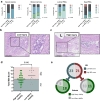
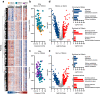
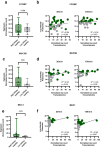
Normothermic machine perfusion (NMP) after static cold storage is increasingly used for preservation and assessment of human donor livers prior to transplantation. Biliary viability assessment during NMP reduces the risk of post-transplant biliary complications. However, understanding of molecular changes in the biliary system during NMP remains incomplete. We performed an in-depth, unbiased proteomics analysis of bile collected during sequential hypothermic machine perfusion, rewarming and NMP of 55 human donor livers. Longitudinal analysis during NMP reveals proteins reflective of cellular damage at early stages, followed by upregulation of secretory and immune response processes. Livers with bile chemistry acceptable for transplantation reveal protein patterns implicated in regenerative processes, including cellular proliferation, compared to livers with inadequate bile chemistry. These findings are reinforced by detection of regenerative gene transcripts in liver tissue before machine perfusion. Our comprehensive bile proteomics and liver transcriptomics data sets provide the potential to further evaluate molecular mechanisms during NMP and refine viability assessment criteria.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

