Salivary microbiome changes distinguish response to chemoradiotherapy in patients with oral cancer
- PMID: 38037123
- PMCID: PMC10687843
- DOI: 10.1186/s40168-023-01677-w
Salivary microbiome changes distinguish response to chemoradiotherapy in patients with oral cancer
Abstract
Background: Oral squamous cell carcinoma (SCC) is associated with oral microbial dysbiosis. In this unique study, we compared pre- to post-treatment salivary microbiome in patients with SCC by 16S rRNA gene sequencing and examined how microbiome changes correlated with the expression of an anti-microbial protein.
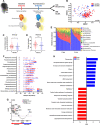
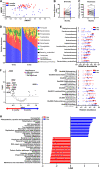
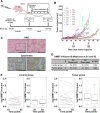
Results: Treatment of SCC was associated with a reduction in overall bacterial richness and diversity. There were significant changes in the microbial community structure, including a decrease in the abundance of Porphyromonaceae and Prevotellaceae and an increase in Lactobacillaceae. There were also significant changes in the microbial community structure before and after treatment with chemoradiotherapy, but not with surgery alone. In patients treated with chemoradiotherapy alone, several bacterial populations were differentially abundant between responders and non-responders before and after therapy. Microbiome changes were associated with a change in the expression of DMBT1, an anti-microbial protein in human saliva. Additionally, we found that salivary DMBT1, which increases after treatment, could serve as a post-treatment salivary biomarker that links to microbial changes. Specifically, post-treatment increases in human salivary DMBT1 correlated with increased abundance of Gemella spp., Pasteurellaceae spp., Lactobacillus spp., and Oribacterium spp. This is the first longitudinal study to investigate treatment-associated changes (chemoradiotherapy and surgery) in the oral microbiome in patients with SCC along with changes in expression of an anti-microbial protein in saliva.
Conclusions: The composition of the oral microbiota may predict treatment responses; salivary DMBT1 may have a role in modulating the oral microbiome in patients with SCC. After completion of treatment, 6 months after diagnosis, patients had a less diverse and less rich oral microbiome. Leptotrichia was a highly prevalent bacteria genus associated with disease. Expression of DMBT1 was higher after treatment and associated with microbiome changes, the most prominent genus being Gemella Video Abstract.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

