The genome sequence of a beetle-killing wasp, Tiphia femorata (Fabricius, 1775)
- PMID: 38037557
- PMCID: PMC10687388
- DOI: 10.12688/wellcomeopenres.18893.1
The genome sequence of a beetle-killing wasp, Tiphia femorata (Fabricius, 1775)
Abstract
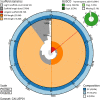
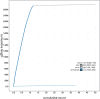
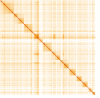
We present a genome assembly from an individual female Tiphia femorata (a beetle-killing wasp; Arthropoda; Insecta; Hymenoptera; Tiphilidae). The genome sequence is 276 megabases in span. Most of the assembly (98.73%) is scaffolded into 12 chromosomal pseudomolecules. The complete mitochondrial genome was also assembled and is 22.4 kilobases in length. Annotation of the genome in Ensembl identified 10,470 protein-coding genes.
Keywords: Hymenoptera; Tiphia femorata; beetle killing wasp; chromosomal; genome sequence.
Copyright: © 2023 Crowley LM et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


References
-
- Archer ME: Tiphia femorata Fabricus,1775.Bees, Wasps & Ants Recording Society,2001. Reference Source
-
- Bogusch P: Vespoidea: Tiphiidae (trněnkovití). Acta Entomologica Musei Nationalis Pragae. 2007;85–92. Reference Source
Grants and funding
LinkOut - more resources
Full Text Sources

