DAXX promotes centromeric stability independently of ATRX by preventing the accumulation of R-loop-induced DNA double-stranded breaks
- PMID: 38038252
- PMCID: PMC10853780
- DOI: 10.1093/nar/gkad1141
DAXX promotes centromeric stability independently of ATRX by preventing the accumulation of R-loop-induced DNA double-stranded breaks
Abstract
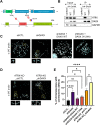
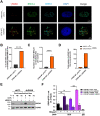
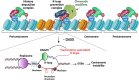
Maintaining chromatin integrity at the repetitive non-coding DNA sequences underlying centromeres is crucial to prevent replicative stress, DNA breaks and genomic instability. The concerted action of transcriptional repressors, chromatin remodelling complexes and epigenetic factors controls transcription and chromatin structure in these regions. The histone chaperone complex ATRX/DAXX is involved in the establishment and maintenance of centromeric chromatin through the deposition of the histone variant H3.3. ATRX and DAXX have also evolved mutually-independent functions in transcription and chromatin dynamics. Here, using paediatric glioma and pancreatic neuroendocrine tumor cell lines, we identify a novel ATRX-independent function for DAXX in promoting genome stability by preventing transcription-associated R-loop accumulation and DNA double-strand break formation at centromeres. This function of DAXX required its interaction with histone H3.3 but was independent of H3.3 deposition and did not reflect a role in the repression of centromeric transcription. DAXX depletion mobilized BRCA1 at centromeres, in line with BRCA1 role in counteracting centromeric R-loop accumulation. Our results provide novel insights into the mechanisms protecting the human genome from chromosomal instability, as well as potential perspectives in the treatment of cancers with DAXX alterations.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Sansregret L., Vanhaesebroeck B., Swanton C.. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018; 15:139–150. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

