Genetic Modification of a Hox Locus Drives Mimetic Color Pattern Variation in a Highly Polymorphic Bumble Bee
- PMID: 38039153
- PMCID: PMC10724181
- DOI: 10.1093/molbev/msad261
Genetic Modification of a Hox Locus Drives Mimetic Color Pattern Variation in a Highly Polymorphic Bumble Bee
Abstract
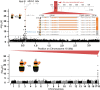
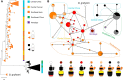
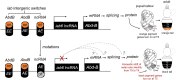
Müllerian mimicry provides natural replicates ideal for exploring mechanisms underlying adaptive phenotypic divergence and convergence, yet the genetic mechanisms underlying mimetic variation remain largely unknown. The current study investigates the genetic basis of mimetic color pattern variation in a highly polymorphic bumble bee, Bombus breviceps (Hymenoptera, Apidae). In South Asia, this species and multiple comimetic species converge onto local Müllerian mimicry patterns by shifting the abdominal setal color from orange to black. Genetic crossing between the orange and black phenotypes suggested the color dimorphism being controlled by a single Mendelian locus, with the orange allele being dominant over black. Genome-wide association suggests that a locus at the intergenic region between 2 abdominal fate-determining Hox genes, abd-A and Abd-B, is associated with the color change. This locus is therefore in the same intergenic region but not the same exact locus as found to drive red black midabdominal variation in a distantly related bumble bee species, Bombus melanopygus. Gene expression analysis and RNA interferences suggest that differential expression of an intergenic long noncoding RNA between abd-A and Abd-B at the onset setal color differentiation may drive the orange black color variation by causing a homeotic shift late in development. Analysis of this same color locus in comimetic species reveals no sequence association with the same color shift, suggesting that mimetic convergence is achieved through distinct genetic routes. Our study establishes Hox regions as genomic hotspots for color pattern evolution in bumble bees and demonstrates how pleiotropic developmental loci can drive adaptive radiations in nature.
Keywords: Hox; Müllerian mimicry; bumble bees; genomic hotspot; noncoding RNA.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of interest statement. All authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

