Hsa-LINC02418/mmu-4930573I07Rik regulated by METTL3 dictates anti-PD-L1 immunotherapeutic efficacy via enhancement of Trim21-mediated PD-L1 ubiquitination
- PMID: 38040417
- PMCID: PMC10693898
- DOI: 10.1136/jitc-2023-007415
Hsa-LINC02418/mmu-4930573I07Rik regulated by METTL3 dictates anti-PD-L1 immunotherapeutic efficacy via enhancement of Trim21-mediated PD-L1 ubiquitination
Abstract
Background: Limited response to programmed death ligand-1 (PD-L1)/programmed death 1 (PD-1) immunotherapy is a major hindrance of checkpoint immunotherapy in non-small cell lung cancer (NSCLC). The abundance of PD-L1 on the tumor cell surface is crucial for the responsiveness of PD-1/PD-L1 immunotherapy. However, the negative control of PD-L1 expression and the physiological significance of the PD-L1 inhibition in NSCLC immunotherapy remain obscure.
Methods: Bioinformatics analysis was performed to profile and investigate the long non-coding RNAs that negatively correlated with PD-L1 expression and positively correlated with CD8+T cell infiltration in NSCLC. Immunofluorescence, in vitro PD-1 binding assay, T cell-induced apoptosis assays and in vivo syngeneic mouse models were used to investigate the functional roles of LINC02418 and mmu-4930573I07Rik in regulating anti-PD-L1 therapeutic efficacy in NSCLC. The molecular mechanism of LINC02418-enhanced PD-L1 downregulation was explored by immunoprecipitation, RNA immunoprecipitation (RIP), and ubiquitination assays. RIP, luciferase reporter, and messenger RNA degradation assays were used to investigate the m6A modification of LINC02418 or mmu-4930573I07Rik expression. Bioinformatics analysis and immunohistochemistry (IHC) verification were performed to determine the significance of LINC02418, PD-L1 expression and CD8+T cell infiltration.
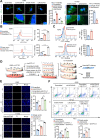
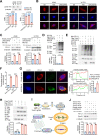
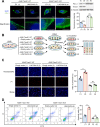
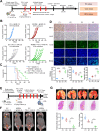
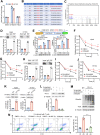
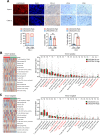
Results: LINC02418 is a negative regulator of PD-L1 expression that positively correlated with CD8+T cell infiltration, predicting favorable clinical outcomes for patients with NSCLC. LINC02418 downregulates PD-L1 expression by enhancing PD-L1 ubiquitination mediated by E3 ligase Trim21. Both hsa-LINC02418 and mmu-4930573I07Rik (its homologous RNA in mice) regulate PD-L1 therapeutic efficacy in NSCLC via Trim21, inducing T cell-induced apoptosis in vitro and in vivo. Furthermore, METTL3 inhibition via N6-methyladenosine (m6A) modification mediated by YTHDF2 reader upregulates hsa-LINC02418 and mmu-4930573I07Rik. In patients with NSCLC, LINC02418 expression is inversely correlated with PD-L1 expression and positively correlated with CD8+T infiltration.
Conclusion: LINC02418 functions as a negative regulator of PD-L1 expression in NSCLC cells by promoting the degradation of PD-L1 through the ubiquitin-proteasome pathway. The expression of LINC02418 is regulated by METTL3/YTHDF2-mediated m6A modification. This study illuminates the underlying mechanisms of PD-L1 negative regulation and presents a promising target for improving the effectiveness of anti-PD-L1 therapy in NSCLC.
Keywords: immune checkpoint inhibitors; lung neoplasms; non-small cell lung cancer; programmed cell death 1 receptor.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
