SMARCB1 loss activates patient-specific distal oncogenic enhancers in malignant rhabdoid tumors
- PMID: 38040699
- PMCID: PMC10692191
- DOI: 10.1038/s41467-023-43498-3
SMARCB1 loss activates patient-specific distal oncogenic enhancers in malignant rhabdoid tumors
Abstract
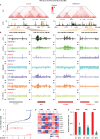
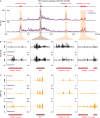
Malignant rhabdoid tumor (MRT) is a highly malignant and often lethal childhood cancer. MRTs are genetically defined by bi-allelic inactivating mutations in SMARCB1, a member of the BRG1/BRM-associated factors (BAF) chromatin remodeling complex. Mutations in BAF complex members are common in human cancer, yet their contribution to tumorigenesis remains in many cases poorly understood. Here, we study derailed regulatory landscapes as a consequence of SMARCB1 loss in the context of MRT. Our multi-omics approach on patient-derived MRT organoids reveals a dramatic reshaping of the regulatory landscape upon SMARCB1 reconstitution. Chromosome conformation capture experiments subsequently reveal patient-specific looping of distal enhancer regions with the promoter of the MYC oncogene. This intertumoral heterogeneity in MYC enhancer utilization is also present in patient MRT tissues as shown by combined single-cell RNA-seq and ATAC-seq. We show that loss of SMARCB1 activates patient-specific epigenetic reprogramming underlying MRT tumorigenesis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- 850571/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 865459/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 338/Stichting Kinderen Kankervrij (KiKa)
- 203.003/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organisation for Scientific Research)
- 181.014/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organisation for Scientific Research)
- 016.16.316/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organisation for Scientific Research)
- P2BSP3-174991/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- LT000209/2018-L/Human Frontier Science Program (HFSP)
- 798573/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

